1. Background
Alpha-1-antitrypsin (AAT) is a glycoprotein that inhibits elastase and is considered as serine protease inhibitors termed serpins. The AAT deficiency causes target organ injury by both loss-of-function and gain-of-toxic function mechanisms. Besides, the accumulation of retained mutant proteins in the endoplasmic reticulum of hepatocytes in a polymerized form may cause chronic liver disease as well as predisposition to hepatocellular carcinoma by gain-of-toxic function (1). AAT deficiency is a common genetic disorder all around the world, which its prevalence varies depending on the population (2) Evidence from studies conducted on the Caucasian population noted AAT deficiency is a major cause of liver disorders in children (3).
2. Objectives
The current study aimed to evaluate the association between specific alleles of AAT with idiopathic neonatal jaundice disease.
3. Methods
3.1. Subjects
The study population consisted of 30 neonates admitted to Ghaem and Dr. Sheikh Hospitals, with a clinical diagnosis of cholestatic idiopathic liver disease. The study also had a control group, which consisted of 30 subjects.
3.2. Methods
Blood samples were taken and stored at 4°C before DNA isolation. The DNA was extracted using the salting out method. After which polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was performed using the following primers:
PFZ: 5´ -GGCTGTGCTGACCATCGTTC- 3´
PRZ: 5´ -AACTCTTCTTTAATGTCATCGAGG- 3´
PFS: 5´ -AGGGGAAACTACAGCACCTCG- 3´
PRS: 5´ -TGGGTACTGTTCTCCTCATCGAGCATG- 3´
The PCR product was digested with Taq I restriction enzyme at 65°C overnight. Subsequently, electrophoresis was performed on a 10% polyacrylamide gel. Direct sequencing was performed on both strands using an ABI Prism BigDye Terminator Cycle Sequencing Kit, version 3.1 (Applied Biosystems) on an ABI Prism 3100 sequencer. All exon PCR fragments were sequenced using the published primers (4).
4. Results
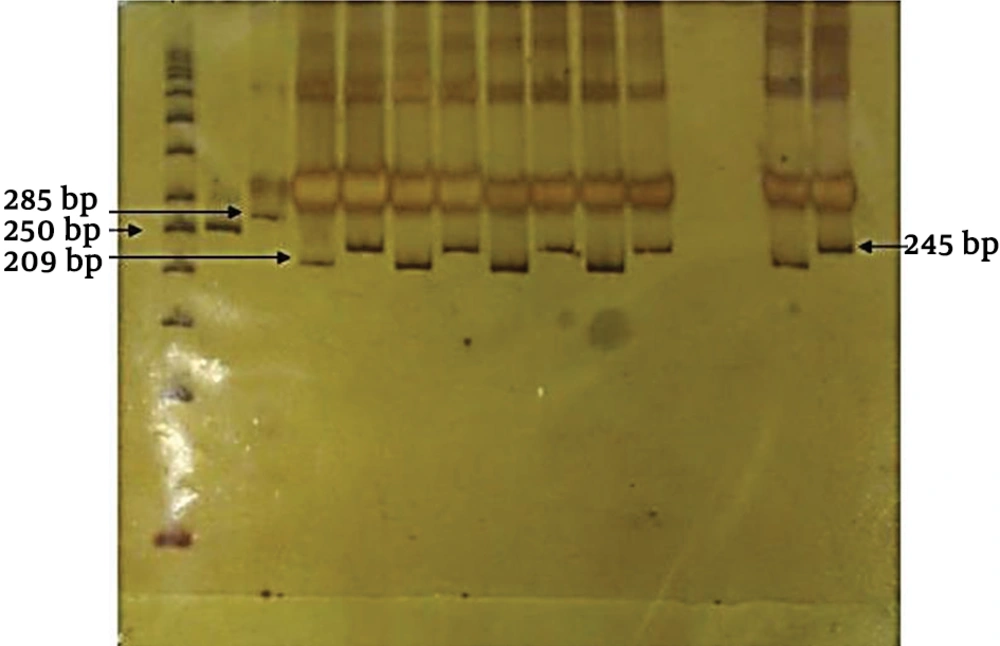
In this study, 30 neonates with prolonged jaundice were investigated. Based on the results of mutant allele using PCR-RFLP method, no case was found with mutant allele. All patients were MM homozygotes (Figure 1).
PCR-RFLP results of patients with MM genotypes. PCR products were digested by Taq I and separated on 10% Polyacrylamide gel electrophoresis (PAGE). (1) 50 bps ladder, (2) Undigested Z allele product (250 bp), (3) Undigested S allele product (285 bp), (4) Non-mutated PCR-Z allele (209 bp), 5) Non-mutated PCR-S allele (245 bp).
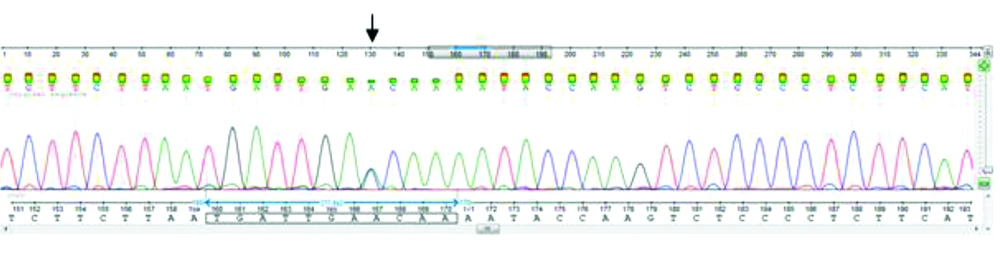
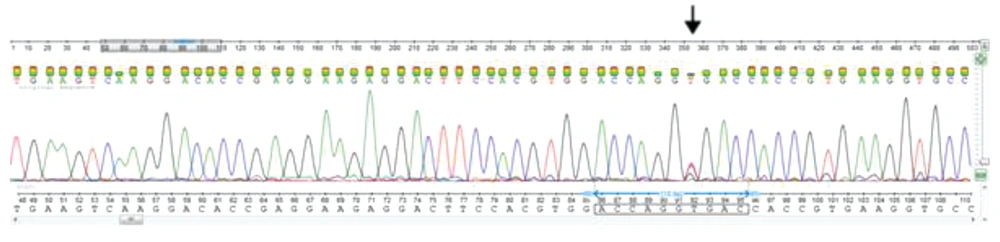
For all 30 patients and those in the control group, exons II, III, IV, and V of the AAT gene were DNA sequenced. Other rare missense variations were detected in exons III, and V. Nucleotide sequence comparison with the AAT sequences available in the GenBank (Accession number, NM_001127705) demonstrated an A to C substitution at position 1200 (Glu400Asp) (Figure 2) and T to C missense mutation at position 710 (Val237Ala) (Figure 3). The substitution of glutamic acid at position 400 by Aspartic acid in exon V of the gene detected in the form of heterozygous (23%), homozygous (10%), and compound heterozygous with p. Val237Ala mutation (10%) in the PiM allele. This variation resulted in replacement by an amino acid of similar nature. Exon III DNA sequencing showed heterozygous variation at position 710 in 10%, and the compound heterozygout mutations p. Val237Ala / p. Asp280Val was observed in one patient. No variation was observed in the controls.
5. Discussion
In this study, the possibility of AAT deficiency was investigated in infants with neonatal cholestasis and/or chronic hepatic disease of undefined etiology. The most common reported deficient alleles associated with liver disease are PiS (Glu264Val) and PiZ (Glu342Lys). The Z allele leads to the formation of a protein that aggregates as periodic acid-Schiff–positive inclusions of AAT within the per portal cells of the liver (5, 6). The S allele is the consequent formation of an unstable protein structure (7). Previous studies reported that the prevalence of AAT deficiency varies in different ethnic groups with liver disease or liver transplantation (8-11). Few studies have investigated the role of AAT in Iranian patients with neonatal jaundice and cholestasis (4, 11, 12). Reports from Asia, over the past years, have reported no AAT deficiency in patients with jaundice (13-15).
In a study conducted in Iran, Lotfi et al. (2005) used the PCR-RFLP and reported a very low incidence of homozygotes in patients with liver and pulmonary diseases (16). There are other studies which confirmed the absence of ATT deficiency in Iranian patients with cholestasis and jaundice (4, 17, 18). Our results are in accordance with previously published data from Iran, mentioning the low prevalence of AAT deficiency in infants with cholestatic idiopathic hepatic disease. Our observations showed other rare variants in the PiM, which might be caused by the AAT deficiency and presenting the neonatal cholestasis. In conclusion, further studies are required to make a conclusion regarding the role of the aforementioned variations on the function of AAT. Also, further studies in different regions of Iran are needed to investigate more variants in ATT, which is common in Iran.