1. Background
In recent years, organ transplantation has become the standard treatment for patients with end-stage organ failure. The survival rates of patients and allografts have been improving due to the development of effective immunosuppressive (IS) agents; however, acute rejection of allografts remains a major obstacle to successful transplantation. Moreover, repeated episodes of acute rejection are considered a major risk factor for developing chronic rejection (1). In addition, the side effects of conventional immunosuppressive drugs, which must be administered for life, necessitate the introduction of alternatives with fewer adverse effects. Current immunosuppressive drugs can disrupt glucose and lipid metabolism, affect bone density, exert neurologic effects, and increase the rates of infection and malignancy in transplant recipients (2). Thus, there is a need to develop more specific drugs for the efficient suppression of alloimmune responses.
A novel category of IS agents that has recently been used to treat refractory forms of autoimmune and allergic disorders is JAK inhibitors (JAKinibs). Janus kinases (JAKs) are enzymes involved in the JAK/signal transducer and activator of transcription (STAT) signaling pathway, which conveys cytokine signals to the nucleus through phosphorylated STAT dimers (3). Therefore, it is presumed that JAK inhibition might modulate immune responses and increase tolerance to the allograft (4). Accordingly, there have been some reports on the efficacy of the JAK-3 inhibitor tofacitinib in preventing renal transplant rejection and improving allograft survival, comparable to cyclosporine, while lower interstitial fibrosis/tubular atrophy (IF/TA) and higher estimated glomerular filtration rates were observed in patients treated with tofacitinib (5, 6).
Despite these findings, there are few studies on the gene expression of JAK enzymes in kidney diseases. For instance, one study has shown elevated tubulointerstitial expression of JAK genes in progressive diabetic nephropathy compared to healthy individuals (7). Further, enhanced JAK/STAT activation, demonstrated by increased levels of transcriptomes, has been reported in the peripheral blood monocytes and biopsy samples of patients with focal segmental glomerular sclerosis (FSGS) (8). However, there is no evidence of JAK expression alterations in kidney allografts.
2. Objectives
Therefore, our aim was to evaluate the gene expression of JAK-1, -2, and -3 in transplanted patients with TCMR compared to stable transplant recipients. In addition to the diagnostic and prognostic value of altered JAK expression, significant upregulation of these genes might suggest them as potential targets for treating TCMR with JAKinibs.
3. Methods
3.1. Patients
Thirty kidney transplant recipients with TCMR, along with 30 age- and sex-matched stable transplant recipients from three medical centers in Tehran, were enrolled in the study between June 2021 and March 2023. Peripheral blood sampling was performed after the diagnosis of TCMR and before the initiation of treatment. The diagnosis of TCMR was made based on either pathological (n = 11) or clinical findings (n = 19) (9). Patients with autoimmunity, allergies, malignancies, or active infections were excluded from the study. All participants provided informed consent. The Ethics Committee of Tehran University of Medical Sciences approved the study (approval code: IR.TUMS.CHMC.REC.1400.005).
3.2. RNA Extraction and CDNA Synthesis
Isolation of ribonucleic acid (RNA) was performed using the High Pure RNA Isolation Kit (ROJE Technologies, Tehran, Iran) according to the manufacturer’s instructions. The quality of the RNA was evaluated with a spectrophotometer (NanoDrop ND1000; Thermo Scientific, USA). RNA samples with an A260/A280 absorbance ratio of 1.8 - 2.2 and an A260/A230 ratio of 2 - 2.2 were considered acceptable. Reverse transcription to complementary deoxyribonucleic acid (cDNA) was performed using the transcriptor first strand cDNA Synthesis Kit (ROJE Technologies, Tehran, Iran). cDNA samples with A260/A280 ratios of 1.8 - 2 were stored in a -70°C freezer for the RT-PCR test.
3.3. Real-time Polymerase Chain Reaction
Relative gene expression was analyzed using SYBR-Green real-time polymerase chain reaction (RT-PCR). Glyceraldehyde 3-phosphate dehydrogenase (GPDH) was used as the internal control, with reverse and forward primer sequences shown in Table 1.
| Gene | Sequence (5’ – 3’) | Company |
|---|---|---|
| JAK1 | ||
| Forward | GAGACAGGTCTCCCACAAACAC | OriGene Technologies |
| Reverse | GTGGTAAGGACATCGCTTTTCC | OriGene Technologies |
| JAK2 | ||
| Forward | CCAGATGGAAACTGTTCGCTCAG | OriGene Technologies |
| Reverse | GAGGTTGGTACATCAGAAACACC | OriGene Technologies |
| JAK3 | ||
| Forward | AGTGACCCTCACTTCCTGCTGT | OriGene Technologies |
| Reverse | GGCTGAACCAAGGATGATGTGG | OriGene Technologies |
| GAPDH | ||
| Forward | GTCTCCTCTGACTTCAACAGCG | OriGene Technologies |
| Reverse | ACCACCCTGTTGCTGTAGCCAA | OriGene Technologies |
To perform the test, 10 μL of master mix (RealQ Plus Green, Ampliqon, Denmark), 7 μL of distilled water, 1 μL of assay mix (forward and reverse primers), and 2 μL of diluted sample cDNA (5 ng/μL) were combined in the wells. The reaction cycles were as follows: 50°C for 2 minutes, 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute, provided by the StepOnePlus real-time PCR System (Applied Biosystems, USA). Nontemplate controls were included in each run. All tests were performed in duplicate and quantified relative to the expression of the internal control. The threshold cycle number was used to calculate the relative expression of the samples, which was determined using the Livak formula: Relative mRNA expression = (2–∆Ct) (10).
3.4. Statistical Analysis
Data analysis was performed using IBM SPSS 26.0 (IBM Corp., Armonk, NY, USA). The data were presented as mean ± standard deviation (SD). The normality of the distribution was evaluated with the Kolmogorov-Smirnov test. An independent samples t-test was used to compare variables with a normal distribution, while the Mann-Whitney U test was applied for non-normal distributions. The correlation between variables was assessed using Pearson's correlation for normal distributions and Spearman's correlation for non-normal distributions. P-values lower than 0.05 were considered significant.
4. Results
4.1. Demographic Findings of the Renal Transplant Recipients
The patients were middle-aged, with an average age of 45 ± 10 years (mean ± SD). There were significant differences in creatinine (P < 0.0001), urea (P = 0.001), and e[glomerular filtration rate (GFR)] (P < 0.0001) between the stable recipients and the patients diagnosed with TCMR. The clinical information of the study population is presented in Table 2.
| Variables | Stable Recipients | TCMR | P-Value |
|---|---|---|---|
| No. | 30 | 30 | |
| Age (y) | 45.9 ± 10.5 | 44.9 ± 11.8 | 0.6 |
| Gender ratio (M/F) | 18/12 | 18/12 | 1 |
| Creatinine | 1.26 ± 0.4 | 2.67 ± 1.2 | < 0.0001 |
| Urea | 32.28 ± 7.9 | 47 ± 14.5 | 0.001 |
| eGFR | 65.04 ± 13.8 | 28.9 ± 10.6 | < 0.0001 |
| TX duration (y) | 4.43 ± 1.9 | 3.45 ± 2 | 0.5 |
| IS protocol | |||
| CsA/MMF/PRED | 7 | 6 | |
| Tac/MMF/PRED | 18 | 16 | |
| AZA/CsA/PRED | 3 | 4 | |
| Tac/Rap/PRED | 2 | 4 |
Abberivitions: IS, immunosuppressive; WBC, white blood cells; CsA, Cyclosporine; TCMR, T cell-mediated rejection; GFR, glomerular filtration rate; TX, transplantation; AZA, azathioprine; Tac, tacrolimus; MMF, mycophenolate; PRED, prednisone; Rap, rapamycin.
a Values are expressed as mean ± SD or No. (%).
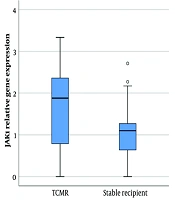
4.2. Increased JAK-1 Expression in TCMR Compared to the Stable Recipients
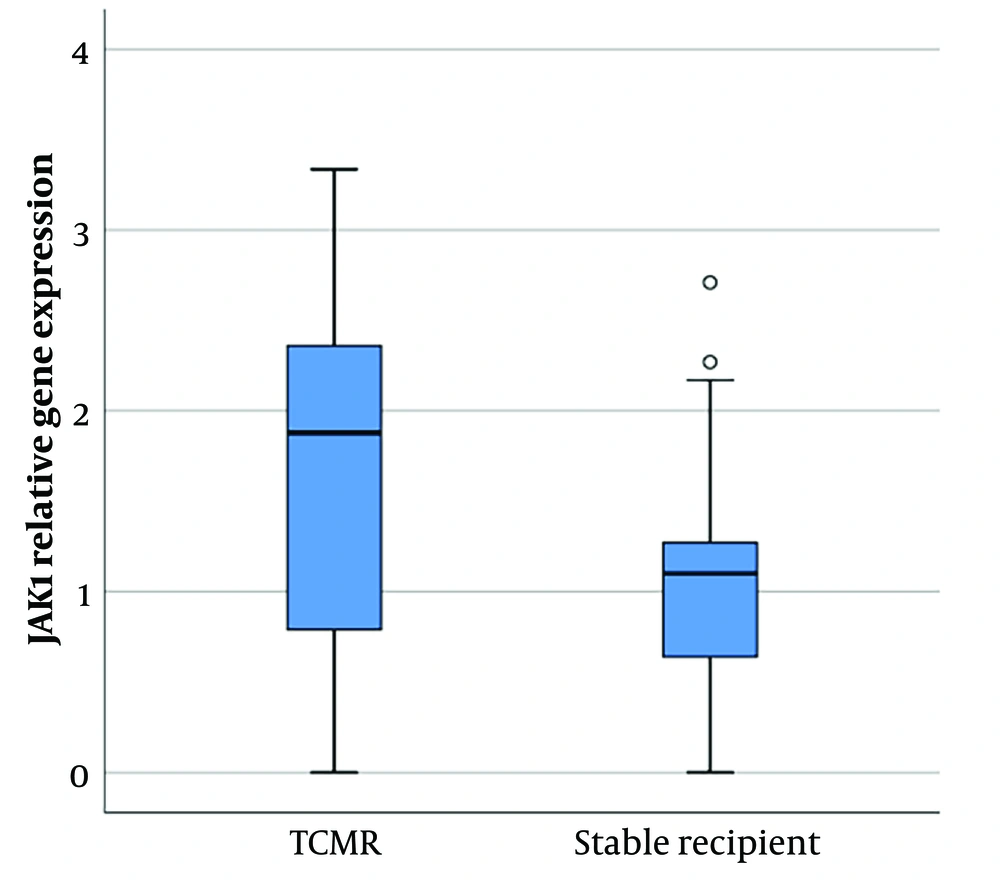
Janus kinase-1 gene expression analysis showed a significant difference between patients with cellular rejection and stable patients (P-value = 0.02). The expression of this gene was significantly higher in patients with TCMR compared to stable recipients (1.64 ± 1.02 vs. 1.07 ± 0.75 [mean ± SD]) (Figure 1).
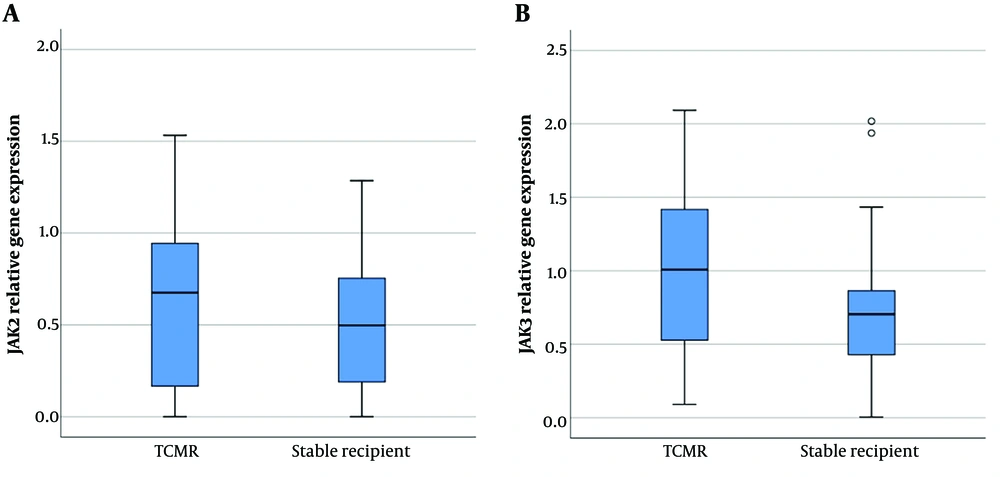
4.3. Comparable mRNA Level of JAK-2 and a Slight Increase in JAK-3 Expression in the TCMR Group
Janus kinase-2 gene expression in PBMCs did not show any significant difference between the two groups (0.63 ± 0.46 vs. 0.51 ± 0.36 [mean ± SD]). Moreover, despite the upregulation of the JAK-3 gene in the TCMR group, the difference did not reach a significant level (0.99 ± 0.57 vs. 0.72 ± 0.47 [mean ± SD]) (Figure 2).
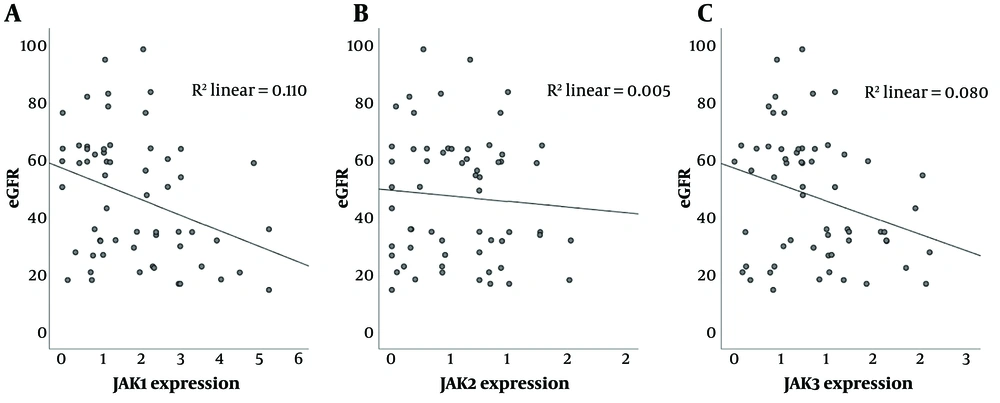
4.4. Negative Correlation Between JAK-1/JAK-3 Expression and eGFR in Kidney Transplant Recipients
Correlation analysis between JAK expression and renal allograft function, as measured by eGFR, demonstrated a significant correlation between JAK-1 and eGFR (correlation coefficient: -0.33, P-value = 0.01) as well as between JAK-3 and eGFR (correlation coefficient: -0.28, P-value = 0.029) in the studied population, including both TCMR and stable patients (Figure 3). However, there was no significant correlation between JAK-2 and glomerular filtration rate.
A, negative correlation between gene expression of Janus kinase (JAK)-1 and glomerular filtration rate (GFR) (P-value = 0.01); B, no significant correlation between JAK-2 gene expression and GFR; C, negative correlation between JAK-3 gene expression and GFR in renal transplant recipients (P-value = 0.029).
5. Discussion
In recent decades, renal transplantation has become a routine therapeutic option for treating patients with end-stage renal disease. The development of advanced surgical methods and novel immunosuppressive drugs has significantly improved organ and patient survival. However, immunologic barriers, including acute, chronic, and antibody-mediated rejection, still affect transplantation outcomes (11). Therefore, several immune components are being studied to identify the most effective targets with the fewest side effects, such as immunodeficiency and metabolic disturbances. To date, the roles of many cytokines and co-stimulatory molecules in allograft rejection, particularly in TCMR, have been studied (12). Our lab has already identified correlations between the gene expression and polymorphism of Toll-like receptor (TLR)-4, TLR-2, Myeloid differentiation primary response 88 (MyD88), Programmed death-ligand 1 (PD-1), Interferon regulatory factor 4 (IRF-4), Helios, Transforming growth factor beta (TGF-β), and interleukins IL-12, IL-15, and IL-18, and T-cell-mediated acute or chronic rejection (13-16).
However, none of these molecules has been considered a promising therapeutic target for treating TCMR, as there are no approved drugs to inhibit these molecules. JAK enzymes, given the growing number of approved JAK inhibitor agents (JAKinibs), appear to be more convenient to suppress compared to other molecules involved in alloimmune responses (17, 18). The benefits of JAKinibs have been demonstrated in a wide range of immune-mediated disorders, such as allergic, rheumatologic, and infectious diseases (19). Since JAK enzymes play a critical role in the signal transduction of cytokines, their inhibition might reduce inflammation in the allograft due to ischemia-reperfusion injury, rejection episodes, and infections such as cytomegalovirus (CMV) (20).
According to this hypothesis, there is a need to evaluate the expression levels of JAK molecules in kidney tissue and peripheral blood in kidney transplant recipients in different states of allograft. Therefore, we aimed to compare the gene expression of JAKs between patients with TCMR and stable renal transplant recipients, which revealed a significant upregulation of JAK-1 and a non-significant elevation of JAK-3 expression in the TCMR group compared to the control patients. Moreover, there was a significant correlation between the expression levels of JAK-1/3 and allograft function as determined by estimated glomerular filtration rates. However, neither the expression assay nor correlation analysis showed a significant difference in JAK-2 expression, but this result might not be conclusive due to the small study population.
These findings suggest that JAK-1 and JAK-3 molecules could be potential targets for managing alloimmune responses in TCMR. Previous findings on the efficacy of tofacitinib, a JAK-3 inhibitor, in preventing acute rejection support these results (5, 6, 21, 22). However, further studies are required to provide compelling data on the efficacy and safety of JAKinibs in the prevention and treatment of TCMR in kidney transplantation. It is also recommended to evaluate the mRNA expression and protein levels of JAKs in renal tissue under different clinical conditions, including rejection, stability, and healthy controls.
5.1. Conclusion
To summarize, the upregulated expression of JAK-1 and JAK-3 genes in the peripheral blood of renal transplant recipients diagnosed with TCMR and the significant correlation between JAK1/3 gene expression and eGFR suggest a probable involvement of the JAK/STAT signaling pathway in rejection. Therefore, JAK inhibitors might be useful in the prevention and treatment of recurrent or refractory forms of TCMR.