1. Background
Gastric cancer (GC) is recognized as the fourth most commonly diagnosed malignancy globally (1, 2). Recent progress in tumor biology and biomarker discovery has driven a transition in cancer treatment from traditional cytotoxic chemotherapy to personalized treatments tailored to the unique characteristics of individual patients, with a focus on the molecular profile of the tumor. Molecular markers have become crucial in the management of gastric carcinoma, serving as critical tools for guiding therapeutic decisions and predicting clinical outcomes (3, 4). Despite this shift, progress in identifying and clinically validating reliable biomarkers for GC has been relatively slow compared to other malignancies (5). Biomarkers, such as nucleic acids, antibodies, peptides, and other molecules, can signify normal physiological states or pathological conditions, including cancer (6). The advent of molecularly targeted therapies has prompted the creation of diverse clinical trial designs focused on identifying patient subgroups with the highest likelihood of responding favorably to these treatments (7). Recent advancements in genome-wide microarray and sequencing technologies have accelerated the identification of novel circular RNAs (circRNAs). Research has shown that the altered expression of circRNAs is linked to the progression of cancer, underscoring their potential as diagnostic markers, including for stomach cancer (8). Furthermore, several studies have proposed that various microRNAs (miRNAs) have the potential to act as useful biomarkers for identifying gastric carcinoma (9, 10). However, several miRNA biomarkers remain unidentified through data analysis utilizing miRNA expression profiling and often lack adequate external validation to confirm their reliability and clinical utility (11-14). Advancements in the development of biomarker-based and molecularly targeted therapies for GC have progressed at a slower pace (15, 16).
2. Objectives
This research seeks to explore the possible involvement of circPVT1 and miR-203a-3p in the early detection of GC. By examining the expression profiles of these markers in cancerous tissues and their nearby non-cancerous counterparts from patients with gastric adenocarcinoma, we seek to assess whether these molecules are implicated in tumor progression.
3. Methods
3.1. Sample Collection and Biomarker Analysis in Gastric Cancer
This observational study assessed the expression of circPVT1 and miR-203a-3p in GC. Twenty-five tumor tissue samples and paired adjacent normal tissues were collected with informed consent and specialist confirmation from patients who underwent surgical resection between 2008 and 2013 at multiple hospitals in Iran. Patients who underwent surgical resection for primary GC at participating hospitals were included in this study. Both the tumor and adjacent non-tumorous tissues were collected. Only cases with complete clinical and pathological information were included in the study, while those with incomplete records were excluded. Samples with poor RNA quality or insufficient tissue quantity were also excluded. Histopathological diagnosis served as the reference standard, and all analyses were conducted blinded to minimize bias. Participant flow and data details are presented in Figure 1 and Table 1. The patient information file (Supplementary File) is included in the appendix. The control group consisted of adjacent normal tissues from the same patients. All samples were rapidly stored in liquid nitrogen and transported under controlled conditions. Both qPCR and histopathological assessments were conducted blinded to minimize bias. Indeterminate qPCR results were retested, and samples with incomplete data were excluded to ensure data integrity. To strengthen the reliability of our results, we utilized data from CircBase and CircInteractome to analyze the expression patterns of these biomarkers in GC. The main goal of this research was to identify biomarkers associated with GC progression. After identifying circRNAs with significant expression changes, circPVT1 was selected for further investigation due to its notable differential expression and limited prior research. The miRNA targets of circPVT1 were identified via CircBank and CircInteractome, leading to the selection of hsa-miR-203a-3p based on a literature review. The expression of hsa-miR-203a-3p was further confirmed using the TCGA dataset through the UALCAN platform.
| Variables | Gender | |
|---|---|---|
| Female (N = 7) | Male (N = 18) | |
| Age | ||
| < 50 | 1 (14.28) | 2 (11.11) |
| 50 - 64 | 1 (14.28) | 1 (5.55) |
| 65 > | 5 (71.42) | 15 (83.33) |
| Tumor stage | ||
| IA | - | 1 (5.55) |
| IB | - | 2 (11.11) |
| II | 1 (14.28) | 4 (22.22) |
| IV | 5 (71.42) | 5 (27.77) |
| IIIA | 1 (14.28) | 6 (33.33) |
| Grade | ||
| 1 | 3 (42.85) | 6 (3.33) |
| 2 | 2 (28.57) | 5 (27.77) |
| 3 | 2 (28.57) | 7 (38.88) |
| TNM (T) b | ||
| T2b | - | 2 (11.11) |
| T3 | 7 (100) | 13 (72.22) |
| T1b | - | 1 (5.55) |
| T2a | - | 2 (11.11) |
| TNM (N) c | ||
| N0 | 1 (14.28) | 9 (50) |
| N1 | 4 (57.14) | 8 (44.44) |
| N2 | 1 (14.28) | - |
| N3 | 1 (14.28) | 1 (5.55) |
| TNM (M) d | ||
| M1 | 4 (57.14) | 4 (22.22) |
| M0 | 3 (42.85) | 14 (77.77) |
a Values are expressed as No. (%).
b T (Tumor): Refers to the dimensions and spread of the initial tumor: T1, the tumor has spread into either the lamina propria (T1a) or the submucosal layer (T1b); T2, cancerous growth has extended into the muscularis propria (T2a) or possibly the submucosa (T2b); T3, the tumor has advanced through to the subserosal tissue.
c N (Nodes): Regional lymph node involvement: N0, no indication of cancer in the surrounding lymph nodes; N1, cancer has reached one or two nearby lymph nodes; N2, three to six regional lymph nodes are affected by cancer; N3, cancer has spread to seven or more nearby lymph nodes, with the extent varying based on specific staging criteria.
d M (Metastasis): M0, there is no indication that the cancer has spread to distant organs; M1, the cancer has extended to distant locations in the body, such as the lungs, liver, or abdominal lining.
3.2. Procedure for Extracting RNA
Total RNA was extracted from gastric tissue samples utilizing the total RNA Extraction Kit (Pars Tous Biotechnology, Iran) based on the RL solution method under RNase-free conditions. Tissue samples were first removed from liquid nitrogen storage and briefly equilibrated at room temperature. Samples were then transferred to a sterile plate and minced thoroughly on ice using a sterile scalpel blade to ensure complete homogenization. All procedures were performed in an RNase-free biosafety cabinet to minimize RNA degradation. Approximately 750 µL of RL solution was added to each 1.5 mL microcentrifuge tube containing the homogenized tissue. The samples were mixed thoroughly and incubated at ambient temperature for 5 minutes. Next, 200 µL of chloroform was introduced, and the tubes were inverted multiple times until the solution turned milky. Phase separation was accomplished through centrifugation at 13,000 rpm for 12 minutes at 4°C. The RNA-containing aqueous phase was carefully transferred to a new RNase-free microtube. To facilitate RNA precipitation, 400 µL of 70% ethanol was added and gently mixed. The resulting mixture was then transferred to a spin column situated in a collection tube, followed by centrifugation at 13,000 rpm for one minute. The flow-through was removed, and the column was rinsed with 700 µL of diluted PW buffer, followed by centrifugation at 13,000 rpm for one minute. To eliminate genomic DNA contamination, 0.5 µL of DNase I enzyme (provided in the Pars Tous kit) was applied directly to the column and incubated at 37°C for 30 minutes. The reaction was terminated by adding one µL of stop solution (containing EDTA). Subsequently, the column was rewashed with 500 µL of PW buffer and centrifuged twice at 13,000 rpm to ensure complete removal of residual contaminants. For the elution step, the column was transferred to a fresh capped microtube, and 35 µL of DEPC-treated water was gently applied to the membrane. After allowing the sample to incubate at room temperature for 3 minutes, it was centrifuged at 13,000 rpm for one minute. The eluted RNA was carefully collected into a 2.0 mL RNase-free microtube, briefly centrifuged, and stored at -70°C for later use.
3.3. Procedure for Synthesizing Complementary DNA
Due to the inherent instability of RNA, reverse transcription was performed to synthesize complementary DNA (cDNA) from total RNA for both mRNA/circRNA and miRNA targets. The total RNA concentration and purity were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). The cDNA targeting hsa_circPVT1 was synthesized using the cDNA Synthesis Kit (Max Cell Biotechnology, Iran). Each reaction mixture included 1000 ng (3 µL) of total RNA, 10 µL of RT buffer, 2 µL of dNTPs, 1 µL of oligo (dT) primer, 1 µL of RT enzyme, and nuclease-free, DEPC-treated water to reach the desired volume. After gentle mixing and brief centrifugation, the reaction was incubated at room temperature for 10 minutes, then heated to 50°C for 1 hour, followed by a 5-minute incubation at 80°C to stop the reaction. For miR-203a-3p, stem-loop reverse transcription was performed using the same cDNA Synthesis Kit (Max Cell Biotechnology, Iran). A solution containing 1 µL of stem-loop primer, 1000 ng (3 µL) of total RNA, and DEPC-treated water was adjusted to a final volume of 7 µL, heated to 70°C for 15 minutes, and immediately cooled on ice for 5 minutes. Next, 10 µL of 2 × RT buffer, 2 µL of dNTP solution, and 1 µL of RT enzyme were added. The reaction was allowed to proceed at 42°C for 1 hour, followed by a final step at 72°C for 5 minutes. The resulting cDNA was stored at -20°C until further use in qRT-PCR analysis.
3.4. Quantitative Assessment of Gene Expression with Real-time PCR
The expression levels of hsa_circPVT1 and hsa-miR-203a-3p were quantified by qRT-PCR in gastric tumor samples and their matched non-cancerous adjacent tissues, which served as the reference standard. To normalize gene expression and account for inter-sample variation, GAPDH was used as the reference gene for circPVT1 quantification, while U6 snRNA served as the internal control for hsa-miR-203a-3p expression analysis. All primers designed for circPVT1 expression were evaluated for target specificity using the NCBI Primer-BLAST tool (Figure 1). The primers were validated based on specificity, GC content, melting temperature (Tm), and the absence of secondary structures or primer-dimer formation. All primers were commercially synthesized, and their sequences are presented in Table 2.
| Gene Names | Seq (5-3) | TM | GC% |
|---|---|---|---|
| circPVT1 F | TTCCTGGTGAAGCATCTG | 54.14 | 50 |
| circPVT1 R | GCACAGCCATCTTGAGG | 55.15 | 58.82 |
| MiR-203a-3p stem loop | GTCGTATCCAGTGCAGGGTCCGAGGTATT; CGCACTGGATCGATACGACCTAGTGG | 79.87 | 56.86 |
| Has-mir-203a-3p | AATCGGCGGTGAAATGTTTAG | 55.92 | 42.85 |
| GAPDH F | AAGGCTGTGGGCAAGGTCATC | 61.78 | 57.14 |
| GAPDH R | GCGTCAAAGGTGGAGGAGTGG | 63.73 | 61.90 |
| U6 F | GCTCGCTTCGGCAGCACATATAC | 64.21 | 56.52 |
| U6 R | CGAATTTGCGTGTCATCCTTGCG | 62.43 | 52.17 |
Abbreviations: Tm, melting temperature; GC, gastric cancer.
3.5. BLAST Alignment Results for circPVT1 Primers
This figure presents the BLAST alignment results of the primers designed for circPVT1 expression using the NCBI Primer-BLAST tool. Both the forward (circPVT1 F) and reverse (circPVT1 R) primers were analyzed to confirm their specificity to the target circPVT1 region. The BLAST analysis parameters included exon junction span (enabled to account for exon-exon boundaries typical of circRNAs), primer specificity check (performed against the RefSeq mRNA database to exclude non-specific amplification), and target size and Tm range (set to optimize binding efficiency and stability), with Tm of 54.14°C for the forward primer and 55.15°C for the reverse primer, as shown in Table 3. The alignment results demonstrate perfect or near-perfect matching of both primers to the circPVT1 sequence with no significant off-target binding. This confirms the suitability of the designed primers for specific and efficient detection of circPVT1 in downstream qPCR assays.
| Primers | Sequence (5´-3´) | Length | Tm | GC% | Self Complementarity | Self 3´ Self Complementarity |
|---|---|---|---|---|---|---|
| Forward | TTCCTGGTGAAGCATCTG | 18 | 54.14 | 50 | 4 | 2 |
| Reverse | GCACAGCCATCTTGAGG | 17 | 55.15 | 58.82 | 2 | 2 |
3.6. Real-time PCR Procedure
Real-time PCR was conducted utilizing SYBR Green chemistry. Each reaction for circPVT1 included 12.5 µL of SYBR Green Real-time PCR Master Mix (Ampliqon, Denmark), primers at a final concentration of 200 nM each, 10.5 µL of DEPC-treated water, and 1000 ng (1 µL) of synthesized cDNA to make a total reaction volume of 25 µL. The PCR cycling protocol began with an initial denaturation step at 95°C for 5 minutes, followed by 40 cycles consisting of denaturation at 95°C for 15 seconds and annealing at 59°C for 30 seconds. For miR-203a-3p quantification, the mixture included 6 µL of SYBR Green Real-time PCR Master Mix (Ampliqon, Denmark), primers at a final concentration of 333 nM each, 4.2 µL of DEPC-treated water, and 1000 ng (1 µL) of cDNA in a total volume of 12 µL. The thermal cycling conditions were identical to those described for circPVT1, except annealing was performed at 50.5°C for 1 minute. All reactions were performed in triplicate, with non-template controls included. Relative expression was determined using the 2-ΔΔCt method. ΔCt represents the difference between the cycle threshold (Ct) of the target and reference genes, and ΔΔCt is calculated as ΔCttumor - ΔCtnormal. Lower ΔCt values indicate higher expression levels. Statistical significance was assessed using GraphPad Prism 10.0. The thermal cycling conditions are listed in Tables 4 and 5.
| Cycle Numbers and Step | Time | Temperature (ᵒC) |
|---|---|---|
| 1 | ||
| Initial denature | 15 min | 95 |
| 2 - 40 | ||
| Denaturation | 15 - 30 s | 95 |
| Annealing | 30 s | 59 |
| Extension | 30 s | 72 |
| Cycle Numbers and Step | Time | Temperature (ᵒC) |
|---|---|---|
| 1 | ||
| Initial denaturation | 15 min | 95 |
| 2 - 40 | ||
| Denaturation | 15 - 30 s | 95 |
| Annealing/extension | 60 s | 50.5 - 60 |
3.7. Data Analysis and Statistical Evaluation
All data analyses were conducted using GraphPad Prism software (version 10). The levels of circPVT1 and miR-203a-3p expression were quantified using the 2-ΔΔCt method, with values expressed as the mean ± standard deviation (SD). A P-value below 0.05 was considered statistically significant. Comparative analysis of expression levels between tumor samples and their corresponding adjacent non-cancerous tissues was performed using paired t-tests (or non-parametric equivalents, if applicable). To compare expression across histological grades (grade I, II, and III), a one-way analysis of variance (ANOVA) was performed on ΔCt values to assess statistically significant differences among the groups.
For evaluating the effects of gender and the interaction between gender and tissue type (tumor vs. normal), mean ΔCt values were analyzed using two-way ANOVA.
3.8. Diagnostic Accuracy Assessment Using ROC Curves
The diagnostic performance of both hsa_circPVT1 and miR-203a-3p in distinguishing GC from corresponding non-tumor tissues was assessed using ROC curve analysis performed in GraphPad Prism (version 10.0). Relative expression levels of each biomarker, quantified through quantitative real-time PCR, were organized in a columnar data table format, with distinct columns representing GC and matched control (normal) tissue samples. ROC analyses were performed by selecting the Analyze function and choosing ROC curve under the Classifier evaluations category in GraphPad Prism.
The software provided key metrics for each biomarker, including the area under the curve (AUC), standard error, 95% confidence interval (CI), and P-value. ROC graphs were generated by plotting sensitivity (%) on the y-axis and (1 - specificity) × 100 on the x-axis. An AUC value close to 1.0 indicated a strong ability to discriminate between groups. A P-value of less than 0.05 was considered statistically significant. Optimal cut-offs from ROC analysis maximized sensitivity and specificity to define test thresholds
4. Results
4.1. Expression Profiling of Circular RNAs and MicroRNAs Related to Gastric Cancer: A Bioinformatics Approach
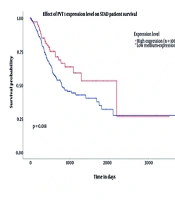
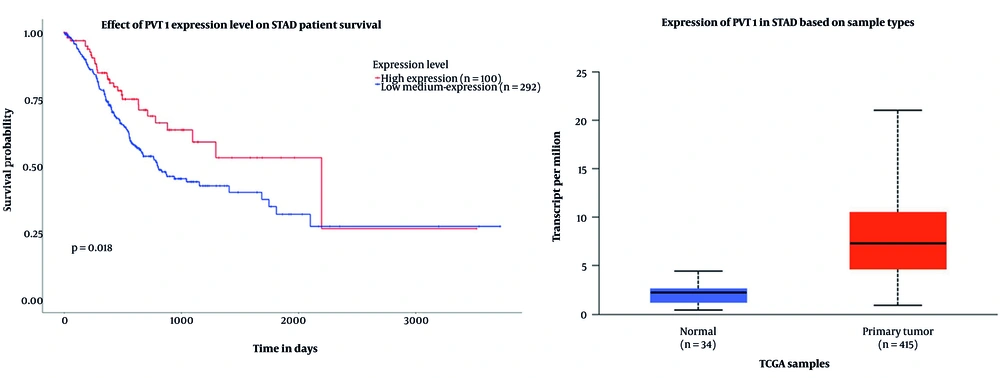
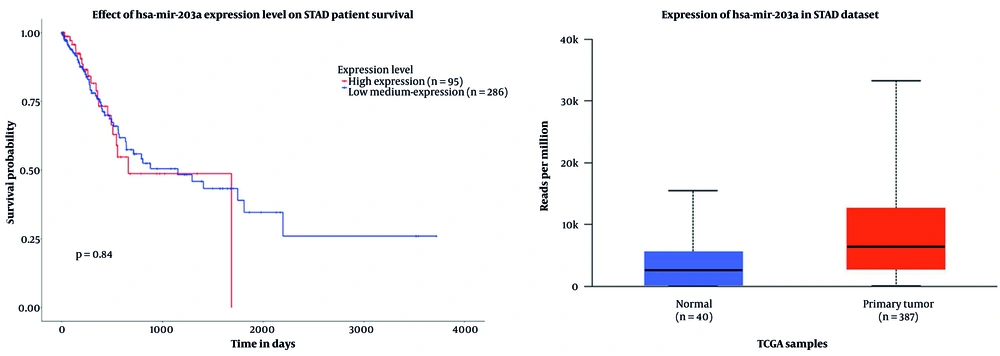
In this study, an initial investigation of circRNAs with altered expression levels was conducted using the CircInteractome database. Among the identified candidates, circPVT1 was selected for further examination, primarily due to the limited existing research on this molecule. Furthermore, analysis using the CircInteractome and CircBank databases suggested that miR-203a-3p may serve as a regulatory target of circPVT1. The bioinformatics analysis focused on the expression profiles of the biomarkers circPVT1 and miR-203a-3p in individuals diagnosed with gastric carcinoma. Data illustrated in Figures 2 and 3 were obtained from the UALCAN platform, emphasizing the expression of miR-203a-3p and the PVT1 gene within the context of GC.
The box plot illustrates the expression levels of the PVT1 gene and hsa-miR-203a-3p in gastric adenocarcinoma (STAD) using TCGA data. For PVT1, the plot shows that higher TPM values correspond to increased gene expression. Normal samples exhibit low PVT1 expression (~ 2 TPM), while primary tumor samples show a significant elevation in PVT1 expression (~ 7 - 8 TPM), indicating that PVT1 is overexpressed in tumor tissues compared to non-tumor tissues. This suggests that PVT1 may play a role in the initiation or progression of gastric carcinoma. The survival curve further indicates that individuals with higher PVT1 expression tend to have better survival outcomes over time compared to those with lower expression levels. While PVT1 is often associated with tumor progression, this observation also implies that other factors, such as treatment response and tumor subtype, may influence survival. Similarly, the box plot for hsa-miR-203a-3p reveals its overexpression in STAD tumors compared to normal gastric tissue, suggesting its involvement in GC development and its potential as a biomarker or therapeutic target. However, survival analysis did not show a significant correlation between hsa-miR-203a-3p expression levels and patient survival outcomes, as the survival curves for high- and low-expression groups were nearly identical. A P-value of 0.84 indicates no significant difference in overall survival based on hsa-miR-203a-3p expression, despite its elevated levels in tumor tissues.
4.2. Expression Analysis of circPVT1 and miR-203a-3p in Gastric Carcinoma Tissue Samples
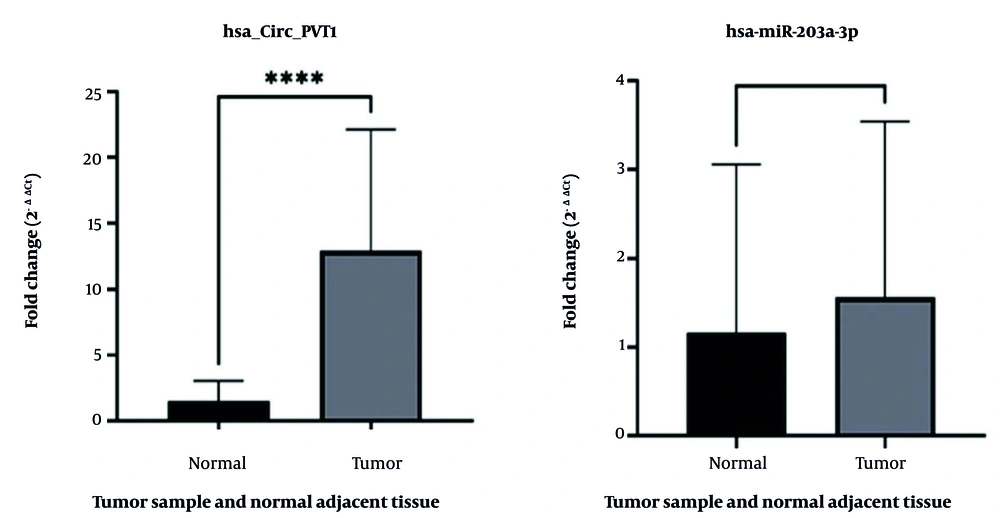
The results demonstrated a significant upregulation of has-circPVT1 expression in tumor tissues compared to normal tissues (P < 0.0001). In contrast, the analysis of hsa-miR-203a-3p expression showed no statistically significant difference between tumor and non-tumor tissues, suggesting that miR-203a-3p may not play a crucial role in the progression of gastric carcinoma or serve as a reliable biomarker for early detection or prognostic evaluation (Figure 4).
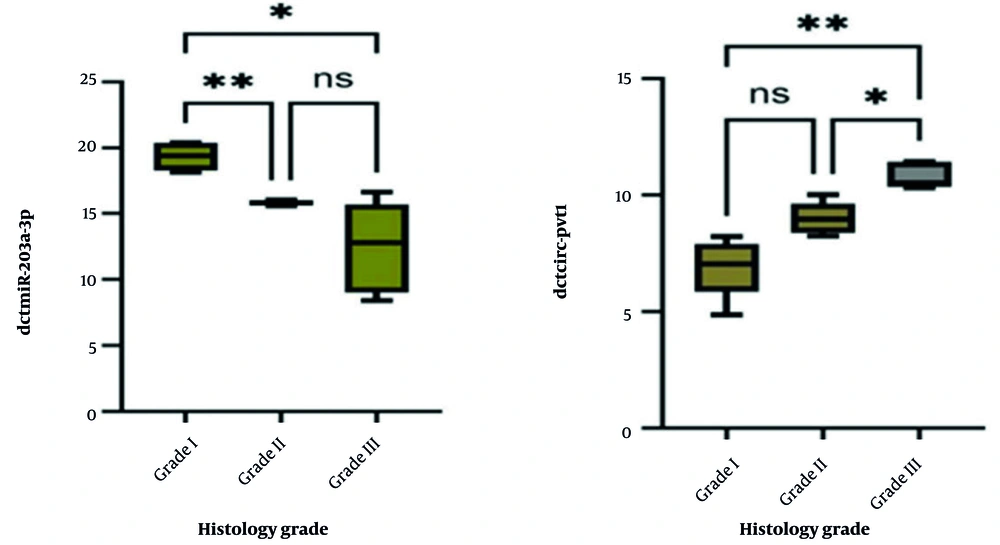
4.3. Expression Profiling of miR-203a-3p and circPVT1 in Gastric Cancer Histological Grades
This part of the study examined variations in the expression of miR-203a-3p and circPVT1 across different histological grades of GC to evaluate their potential roles in disease progression. While the overall comparison did not reveal statistically significant differences across all cancer grades (P > 0.05), a notable association was found between miR-203a-3p expression and tumor grading (P = 0.013). Specifically, expression levels of miR-203a-3p decreased significantly from grade I to both grade II and III, though the difference between grades II and III was not significant. In contrast, circPVT1 expression showed a strong association with cancer grade (P = 0.002), with a significant increase observed from grade I to grade III and from grade II to grade III. No significant change was observed when comparing grade I to grade II. Taken together, these results suggest that both miR-203a-3p and circPVT1 may play a role in the progression of GC, particularly due to the substantial variations in circPVT1 expression across different tumor grades (Figure 5).
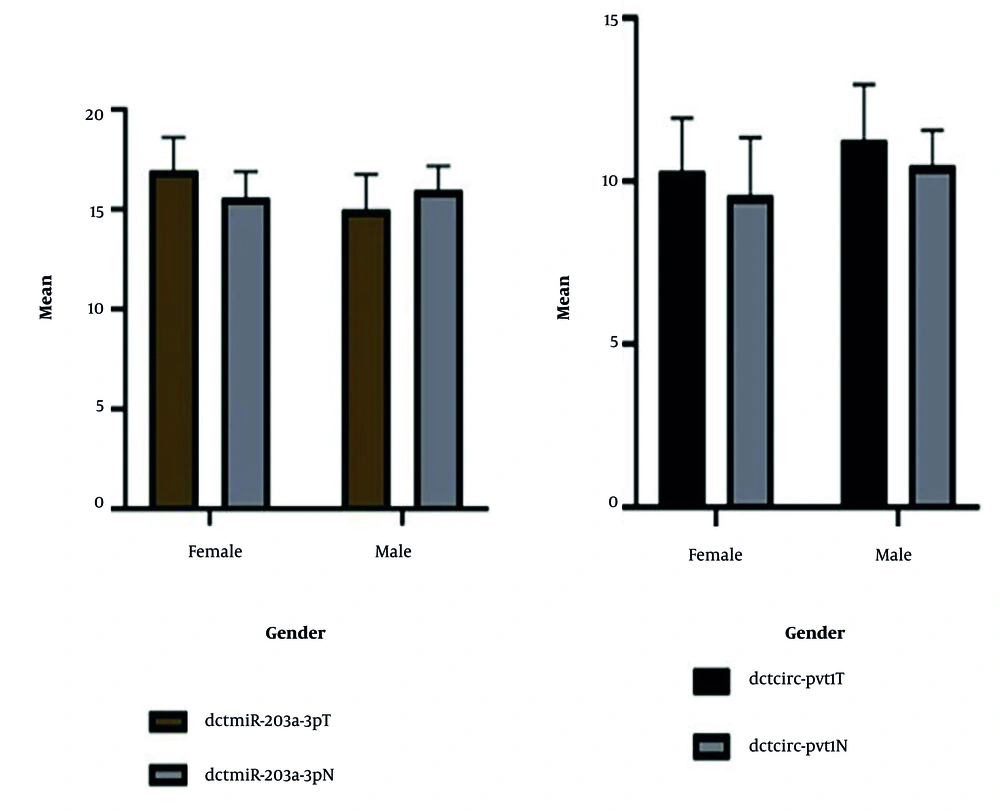
4.4. Differential Expression of miR-203a-3p and circPVT1 by Gender in Normal and Tumor Tissues
The average expression levels of miR-203a-3p and circPVT1 were compared between normal and tumor groups, stratified by gender. Expression plots in Figure 6 revealed no significant differences in miR-203a-3p or circPVT1 expression between male and female patients in both the normal and tumor groups. These results suggest that gender does not significantly impact the expression of miR-203a-3p and circPVT1, indicating that gender may not play a key role in the variability of these biomarkers in gastric carcinoma.
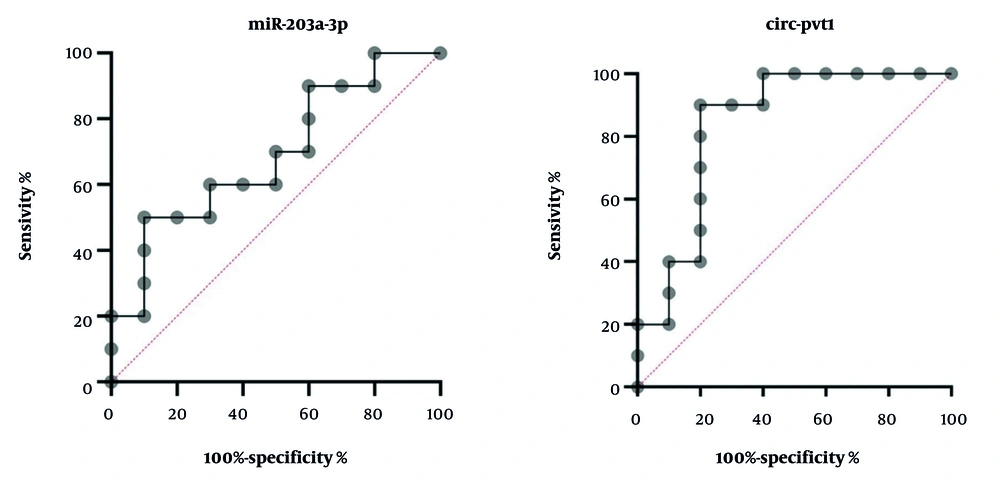
4.5. Evaluation of the Receiver Operating Characteristic Curve
To assess the diagnostic performance of the biomarkers under investigation, we conducted an ROC curve analysis using GraphPad Prism version 10 (Figure 7). For has-circPVT1, the AUC was 0.84, indicating strong diagnostic accuracy. The standard error was 0.096, with a 95% CI ranging from 0.6506 to 1.0000. The P-value was 0.0102, reflecting a significant result (P < 0.05). For miR-203a-3p, the AUC was 0.69, showing a limited ability to discriminate. The standard error was 0.1206, with the 95% CI spanning from 0.4537 to 0.9263. The P-value for miR-203a-3p was 0.1509, indicating no significant effect (P > 0.05).
5. Discussion
Gastric carcinoma remains a significant global health challenge, primarily due to its high prevalence and associated mortality. With the advancement of precision medicine, understanding the molecular mechanisms underlying GC is crucial for enhancing diagnostic accuracy and therapeutic outcomes (17). Jiang et al. identified several genes with altered expression in GC tissues, providing insight into the critical biological processes that drive disease progression (18). circPVT1 has been shown to interact with specific miRNAs, acting as a molecular sponge to inhibit their activity (19, 20). In this study, we combined computational analysis with experimental validation to evaluate the expression profiles and potential clinical relevance of circPVT1 and miR-203a-3p in GC. Our results revealed a significant upregulation of circPVT1 in gastric tumor tissues compared to adjacent non-cancerous tissues (P < 0.0001), consistent with previous studies suggesting its role as a tumor-promoting factor in various cancer types (21, 22). Furthermore, Kun-Peng et al. (as cited by Adhikary et al.) observed an increase in circPVT1 levels in osteosarcoma (OS) samples compared to adjacent normal tissues, suggesting its potential oncogenic role. Their findings indicated that circPVT1 may suppress cell proliferation while increasing the sensitivity of OS cell lines to chemotherapy (23). Additionally, circPVT1 promotes tumor development by regulating the epithelial-mesenchymal transition (EMT), which may drive tumor cell proliferation, invasiveness, and metastasis. In line with this, Hua and Luo found elevated circPVT1 expression in thyroid carcinoma, pointing to its involvement in tumor progression (24). Moreover, research by Li et al. (cited by Shi J.) showed that the transcription factor c-Fos regulates circPVT1 expression, playing a key role in the initiation and progression of non-small cell lung cancer (NSCLC) (25). In 1991, Nigro et al. were the first to suggest that circRNAs could function as potential tumor suppressor transcripts. Subsequently, another study conducted a comprehensive analysis of RNA translation mechanisms in cardiac tissues, identifying several translation products derived from circRNAs (26). Notably, circPVT1 expression was significantly higher in advanced pathological grades of GC. Significant differences were observed when comparing grade I to grade III, as well as grade II to grade III, emphasizing its strong association with tumor progression and aggressiveness. These findings suggest that circPVT1 could be an important marker for assessing disease stage and predicting patient prognosis. Although circPVT1 is commonly recognized as a molecular sponge for miRNAs, particularly those with tumor-suppressive functions such as miR-125 and miR-203a (27), our findings challenge this prevailing model. Specifically, we observed no significant variation in miR-203a-3p expression between tumor tissues and adjacent non-cancerous tissues. This observation contrasts with data from TCGA, which indicate elevated miR-203a-3p levels in gastric carcinoma samples, and with the findings of Yang et al. (as cited by Zu et al.) (28), who identified miR-203a as a tumor suppressor through E2F3 inhibition. Interestingly, we observed a notable increase in miR-203a-3p expression in higher tumor grades (P = 0.013), suggesting its potential role in disease progression rather than initiating tumorigenesis. These discrepancies could arise from differences in sample origin, ethnic background, cancer subtype heterogeneity, or post-transcriptional regulation. In our gender-based analysis, no significant differences were found in the expression of circPVT1 or miR-203a-3p, indicating that these biomarkers may be consistently expressed across various patient groups. To our knowledge, this is one of the first studies to experimentally distinguish circPVT1 overexpression from miR-203a-3p downregulation in GC, suggesting the involvement of alternative mechanisms or additional miRNA targets in circPVT1’s oncogenic role. However, this study was limited by the absence of functional validation (e.g., inhibition or overexpression assays) to confirm direct regulatory interactions. Additionally, our conclusions are partially based on bioinformatic predictions, necessitating further molecular and mechanistic studies. In summary, our findings highlight circPVT1 as a promising biomarker linked to tumor severity and progression in gastrointestinal cancers. Although miR-203a-3p may hold diagnostic value, its role appears to be context-dependent and warrants further investigation.
5.1. Conclusions
This study identifies circPVT1 as a promising circRNA biomarker, exhibiting significantly elevated expression levels in GC tissues. Its strong association with higher tumor grades suggests its utility not only as a diagnostic tool but also as a prognostic marker linked to disease progression. The evidence indicates that circPVT1 may contribute to tumorigenesis by influencing key oncogenic pathways, potentially involving interactions with YAP1, mutations in p53, and suppression of miR-203a-3p. Collectively, these findings enhance our understanding of the role of non-coding RNAs in the development and progression of gastric tumors and provide a strong basis for future research into circRNA-based diagnostics and targeted therapies.