1. Background
Cancer remains a major cause of mortality in both industrialized and many developing nations, resulting in around 10 million fatalities globally (1). Colorectal cancer (CRC) is the third most common type of cancer and the second leading cause of cancer-related mortality, accounting for approximately 10% of all cancer deaths globally (2). The pathogenesis of CRC is a multifaceted phenomenon characterized by the interaction of genetic predispositions and environmental influences, which complicates both prevention and therapeutic approaches (3). Although traditional treatment modalities, including surgery, radiotherapy, and chemotherapy, have demonstrated some degree of efficacy (4), they face considerable challenges such as recurrence, metastasis, and the development of drug resistance (5, 6). Furthermore, the inherent heterogeneity among cancer cells, along with variations among individual patients, adds another layer of complexity to the formulation of effective treatment strategies (7, 8).
Molecular genetics is essential for comprehending CRC, as emphasized by Piawah and Venook (9). This area encompasses the examination of genetic variations, gene mutations, epigenetic modifications, and the regulation of gene expression, which aids in uncovering the intricate mechanisms that play a role in the progression of the disease (10). This type of research is crucial for the identification of potential pathogenic genes, driver mutations, and cancer-associated signaling pathways, thereby offering significant insights for personalized therapies and the development of targeted treatments (11). Genomic investigations have identified several genes associated with the initiation and progression of CRC, including PIK3CA, KRAS, APC, SMAD4, and BRAF, with particular emphasis on the PIK3CA gene (12).
The PIK3CA gene, responsible for encoding the alpha catalytic subunit of phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), is often modified in numerous types of cancer. It is situated on the long arm of human chromosome 3 at the 3q26.32 locus and produces a catalytic protein known as p110, consisting of 1,068 amino acids and having a molecular weight of approximately 110 kDa. The PI3K plays a crucial role in the phosphorylation of phosphatidylinositol-4,5-bisphosphate to form phosphatidylinositol-3,4,5-triphosphate. Mutations in the PIK3CA gene are commonly observed across a diverse array of cancers, particularly in prevalent types such as breast, endometrial, and CRCs. The high frequency of these mutations may offer potential therapeutic strategies, even for less common cancers that are generally challenging to treat (13). Mutations in the PIK3CA gene are significant not only in the progression of CRC but also in shaping its clinical features, prognosis, and treatment responses (14, 15). The E542K and E545K mutations were prioritized due to their high prevalence in global CRC studies and their established role in PI3K pathway activation (13).
Alpelisib was initially discovered in 2013 and has been approved for the treatment of PIK3CA-mutant breast cancer. It is recognized as the first pharmacological inhibitor of the p110α subunit to receive marketing authorization. The widespread presence of PI3Kα in the body can lead to anticipated adverse events when it is inhibited. Notably, alpelisib's interference with insulin signaling may cause glucose intolerance or diabetes, as shown by dose-dependent effects observed in breast cancer patients during the SOLAR study (16, 17).
Emerging inhibitors targeting p110α variants aim to enhance treatment efficacy for mutant cells while improving safety. Notably, LOXO-783, an allosteric small-molecule inhibitor, exhibits high selectivity for the p.H1047R variant of PI3Kα, demonstrating preclinical activity without affecting the wild-type PI3Kα in breast cancer patients (PIKASSO-01, NCT05307705) (18). Additionally, two chemically distinct pan-mutant PI3Kα inhibitors, RLY-2608 and RLY-5836, have shown effectiveness both in vitro and in vivo. They exhibit favorable safety profiles and robust inhibition of mutant PI3Kα (19). Currently, both medications are being evaluated in phase 1 clinical trials for advanced breast cancer (clinical trials NCT05216432 and NCT05759949).
Therefore, it is essential to explore the function and mechanisms of PIK3CA gene mutations in CRC to gain a deeper insight into the molecular pathology of this condition. Such research offers essential insights and direction for early detection, molecular categorization, risk assessment, targeted treatment, and immunotherapy approaches for CRC (12).
2. Objectives
This study aimed to evaluate mutations in the PIK3CA gene, specifically E542K and E545K, linked to CRC in the Iranian population.
3. Methods
3.1. Ethics Statement
This study is approved under the ethical approval code of the Committee of Dezful University of Medical Sciences in Dezful, Iran (code: IR.DUMS.REC.1401.104).
3.2. Patients and Sampling
We identified 39 patients diagnosed with CRC based on histological evaluations and clinical criteria, as confirmed by a pathologist. These patients were selected from referrals to Ganjavian Hospital and Imam Ali Clinic in Khuzestan province, Iran, between January 2015 and April 2022. Tumor tissue samples from these patients were formalin-fixed, paraffin-embedded (FFPE), and sectioned into 5 - 10 µm slices for DNA extraction. We employed the phenol-chloroform method for digestion, followed by ethanol precipitation. Informed consent was obtained from all participants in the study or their family members.
3.3. Genotyping by Amplification Refractory Mutation System Polymerase Chain Reaction
We selected two specific mutations in the PIK3CA gene — E542K (G>A) and E545K (G>A) — as potential hotspot mutations associated with CRC. To detect these mutations, we employed the BIO-RAD T100™ Thermal Cycler System using the amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) method. The PCR reactions were conducted in a total volume of 20 µL, which included 10 µL of Taq DNA Polymerase Master Mix RED, 2 µL of primers, 2 µL of DNA, and 6 µL of double-distilled water (ddH2O). The amplification cycle began with an initial denaturation at 95°C for five minutes. This was followed by 34 cycles consisting of denaturation at 95°C for 30 seconds, annealing at 59°C for E542K or 60°C for E545K for 30 seconds, and extension at 72°C for 20 seconds. The final steps included a single extension cycle at 72°C for five minutes. The primers utilized in this study are detailed in Table 1.
| Gene | Forward Primers | Reverse Primers | Product Length (bp) |
|---|---|---|---|
| E542K G>A | 5’ ATCATGTGTGAATCCAGAGG 3’ | R-Allele G: 5’TTCTCCTGCTCAGTGATTTC3’; R-Allele A: 5’TTCTCCTGCTCAGTGATTTT3’ | 173 |
| E545K G>A | 5’ATCATGTGTGAATCCAGAGG3’ | R-Allele G: 5’TGACAATCTTTCTCCTGCTC3’; R-Allele A: 5’TGACAATCTTTCTCCTGCTT3’ | 182 |
3.4. DNA Sequencing
To validate the findings from ARMS-PCR, we examined the DNA sequencing for the presence of the hotspot mutations E542K G>A and E545K G>A in a randomly selected 20% of the samples. Sequencing was conducted on the ABI 3130XL capillary sequencing platform provided by Applied Biosystems/Life Technologies in Carlsbad, CA, USA. The resulting sequencing data were then analyzed using Chromas version 2.6.6 software.
3.5. Statistical Methods
Associations were analyzed using Fisher’s exact test and independent t-tests via SPSS v26.
4. Results
4.1. Epidemiological and Clinicopathologic Results
In this study, we analyzed 39 patients, comprising 14 women (35.9%) and 25 men (64.1%). The average age of the participants was 59.2 ± 15.9 years. When categorizing tumor sizes, we found that 46.1% of the patients had tumors larger than 4.5 cm, while 53.9% had tumors measuring 4.5 cm or smaller. Lymphoid metastasis was observed in 55.81% of all patients. The incidence of lymphoid metastasis did not significantly differ between women (58.2%) and men (41.8%), with a P-value of 0.717. However, a direct correlation was identified between lymphoid metastasis and tumor size; patients with tumors exceeding 4.5 cm exhibited a notably higher incidence of lymphoid metastasis (P < 0.001).
According to the disease grading system, grades 1 and 2 were the most common among patients. Although individuals with grade 3 had a higher average age, this difference was not statistically significant (P = 0.924). Additionally, there was no significant correlation between tumor size and gender (P = 0.979), age (P = 0.116), or disease grade (P = 0.058). In terms of disease stage, 18.42% of patients were classified as stage 1, 50% as stage 2, and 31.58% as stage 3. Furthermore, no significant associations were found between disease stage and either gender (P = 0.478) or age (P = 0.180).
4.2. Amplification Refractory Mutation System-Polymerase Chain Reaction and Sequence Analysis Results
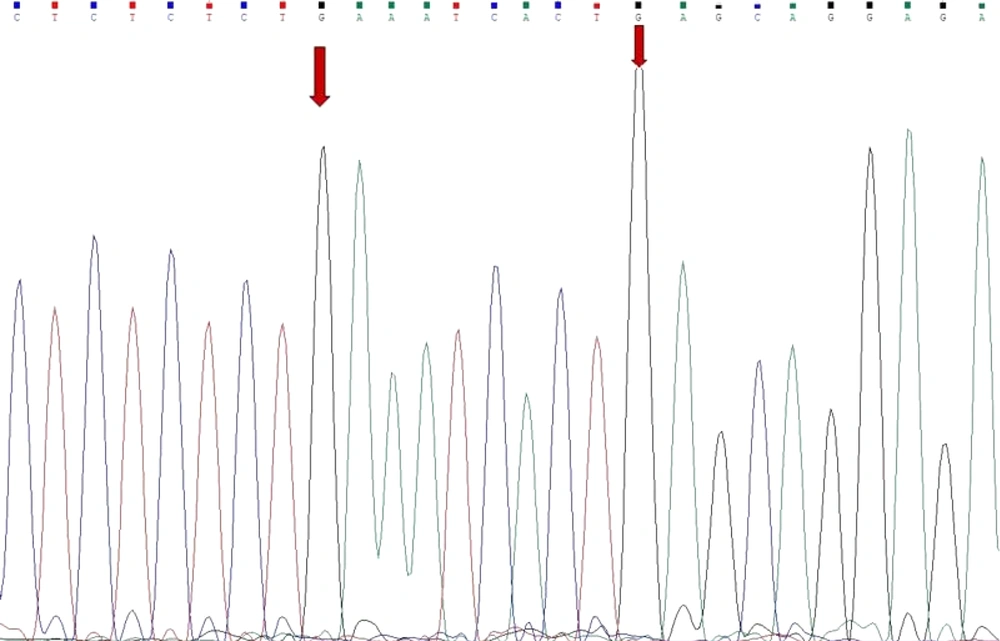
This study utilized the ARMS-PCR technique to analyze mutations. For each mutation at the E542K and E545K loci, the PCR was performed twice: Initially with forward and reverse primers targeting the wild-type allele, and subsequently with forward and reverse primers for the mutant allele. Sequencing of the PCR products was carried out after the PCR was completed. The findings indicated an absence of mutations at the E542K and E545K loci of the PIK3CA gene in patients diagnosed with CRC (Figure 1).
5. Discussion
The CRC remains a significant health challenge, and our research focuses on its association with genetic mutations, particularly those involving the PIK3CA gene. As the third most common cancer and a leading cause of cancer-related deaths worldwide, it is essential to understand the complexities of CRC. Our analysis of 39 tumor samples collected from patients at Dr. Ganjavian Hospital and Imam Ali Clinic in Dezful offers valuable insights into the characteristics of this disease within our local context. Our sample has an average age of just over 59, representing a significant portion of the population at higher risk for CRC. The gender distribution indicates that men are more frequently affected, consistent with global trends (20).
Notably, the tumor size data is alarming: More than 46.1% of patients had tumors larger than 4.5 cm. This raises concerns since larger tumors are often related to more aggressive forms of the disease and poorer outcomes. Our results are consistent with the study by Weixing Dai et al., which demonstrated that tumor size serves as an independent variable influencing overall survival (OS) and disease-free survival (DFS) in individuals diagnosed with colorectal adenocarcinoma (21). One of the key findings from our research is the significant occurrence of lymphoid metastasis — more than half of our patients experienced this complication. Our research aligns with the findings of Kamilla Maria Bech Johannesen et al. (22). The link between larger tumor size and a higher rate of lymphoid metastasis highlights the importance of early detection and intervention (23). Our analysis of tumor grades revealed that most tumors were classified as grades 1 and 2, consistent with the findings of Ueno et al. (24). While 18.42% of patients were diagnosed at stage 1, the fact that 31.58% were at stage 3 suggests that a significant number were diagnosed after the cancer had progressed. This highlights the urgent need for improved screening practices.
Interestingly, our study found no significant associations between disease stage and gender or age, contrasting with some previous reports. For instance, Gabriel et al. observed a correlation between advanced age and higher disease stages (25). This discrepancy could be attributed to our comparatively limited sample size or other factors specific to the population.
Although PIK3CA mutations play a critical role in CRC, our study did not find any evidence of mutations at the E542K and E545K loci in the patients we studied. This finding was unexpected, especially considering the existing literature that frequently identifies these mutations as important contributors to CRC development (26). The absence of these specific mutations in our study may indicate that population-specific genetic variations or environmental factors contribute to CRC, which merits further investigation.
Although we did not find a direct correlation between specific PIK3CA mutations and the presence of CRC in our study, we detected a significant relationship between tumor size and lymphatic metastasis. This finding underscores the importance of ongoing research in this area. Gaining a deeper understanding of the unique genetic landscape of CRC across different populations could lead to more targeted therapies and ultimately improve patient outcomes.
5.1. Conclusions
This study involving 39 CRC patients found no mutations at the PIK3CA loci E542K and E545K in the studied population. The small sample size (n = 39) and focus on only two hotspot mutations (E542K/E545K) may limit the generalizability of our findings. Future studies should include exon 20 (H1047R) and larger multicenter cohorts. The absence of PIK3CA mutations in this cohort suggests that anti-EGFR therapies (e.g., cetuximab) may remain viable options for Iranian CRC patients, as PIK3CA mutations often confer resistance to these treatments. However, a significant association was detected between larger tumor size and an increase in lymphoid metastasis. These findings underscore the importance of population-specific genetic profiling in CRC, even in the absence of expected mutations. Further research should explore PI3K/AKT/mTOR inhibitors in PIK3CA-wild-type CRC patients.