1. Context
Comprehensive analysis of cancer transcriptomes revealed that despite the critical role of mutations in tumorigenesis, perturbation caused by epigenetic and post-transcriptional modifications contributes to cancer progression (1). RNA editing is a pivotal post-transcriptional and/or co-transcriptional modification essential for the development of normal cells and is involved in a wide range of biological processes (2). Editing events are mediated through adenosine deaminases acting on RNA (ADARs) on double-stranded RNA and expanding the transcriptome's diversity and the range of functions of RNA transcripts (3).
Theoretically, editing is capable of changing any nucleotide to another type. In humans, the most frequent type of RNA editing event is adenosine deamination to inosine, which is accomplished by ADARs proteins (A to I deamination) (4, 5). The resulting inosine is interpreted as guanosine (A to G conversion) by cell machinery and acts as a single nucleotide mutation (6). Editing in the coding region may occur with a nonsynonymous effect. On the other hand, editing in non-coding sequences such as microRNA and 3′UTR of mRNAs is more frequent than in coding regions (7, 8). RNA editing in these sequences provides the ability of time-specific and location-specific regulation that changes cellular properties (9). Altogether, balanced and regulated RNA editing is essential for the normal function of cells, while aberrant and dysregulation of this phenomenon may deviate the fate of a normal cell (10).
2. Evidence Acquisition
Science Direct and PubMed databases were reviewed from 2000 to 2020 and 2003 to 2020, respectively. These databases searched using "RNA editing" and "cancer" keywords. The current study aimed to review the role of RNA editing in cancer.
3. Results
3.1. Editing Enzymes
A to G editing is mediated through adenosine deaminases acting on the RNA (ADAR) family of enzymes. Human cells express three kinds of ADARs: ADAR1, ADAR2, and ADAR3 (11). ADAR1 and ADAR2 form homodimers, whereas ADAR3 does not form dimers. The first two are catalytically active enzymes and ubiquitously expressed, while the latest is mainly expressed in the brain (12, 13). ADAR1 is expressed as two isoforms, ADAR1p110, and ADAR1p150. ADAR1p110 edits dsRNA is in the nucleus, while ADAR1p150 editing is in the nucleus and cytoplasm. Expression of ADAR1 is essential, and ADAR1-null mice showed substantial overexpression of interferon and widespread apoptosis. In comparison, ADAR2-null mice live somewhat longer than ADAR1 knockouts and are prone to seizures and die young (14). The structure and sequence of ADAR3 are similar to those of ADAR1 and ADAR2. They share the same domains, such as nuclear localization, deaminase, and RNA-binding (15). It has been reported that ADAR3 may instead act as a dominant-negative form of other ADARs (16).
3.2. Identification of Editing Sites
The development of next-generation sequencing (NGS) technologies and open access data sharing in genomic research have provided inimitable opportunities for genome-wide investigation of RNA editing sites (17). There are two main strategies to identify RNA editing sites using NGS technology. The straightforward but more expensive approach to discover RNA editing events requires simultaneous genome and transcriptome sequencing in the same sample (18). The indirect strategy compares transcriptome sequence with the reference genome, therefore, it needs RNA-Seq data alone, and it is more challenging (19). Both approaches meet several challenges to the accurate identification of RNA editing sites. Discrimination of bona fide editing events from technical noise such as sequencing errors and incorrect mapping is one of the main drawbacks. On the other hand, biological noise such as genomic polymorphisms and somatic mutations may also be interpreted as editing sites (20). Alongside the employment of NGS, researchers have developed another technique called inosine chemical erasing (ICE). ICE is designed for genome-wide profiling of A-to-I editing sites through reverse transcription, PCR amplification, and direct sequencing without comparing the sequence with cDNA or reference genome (21).
3.3. Functional Impacts of Editing Sites
In recent years, a growing body of evidence has revealed that RNA editing events are an important contributor to cancer pathogenesis. Given the location of editing found in the human transcriptome, these editing sites could have different functional impacts on tumorigenesis (22).
3.3.1. RNA Editing Role in the Pathogenesis of Cancer
Tumors benefit from editing sites via several mechanisms, most conveniently through nonsynonymous codon changes (Figure 1). In this regard, antizyme inhibitor 1 (AZIN1) is one of the well-studied genes. A to G editing leads to a serine to glycine conversion at residue 367 in AZIN1, which confers a gain-of-function phenotype (23). Edited AZIN1 promotes stemness and appears to drive the metastatic progression in colorectal cancer (24). It is also reported that activation of AZIN1 RNA editing increases the invasive potential of cancer-associated fibroblasts in colorectal cancer (25). The enhanced rate of AZIN1 editing in gastric cancer is an independent prognostic factor for poor overall survival and significantly correlated with the presence of lymph node metastasis (26).
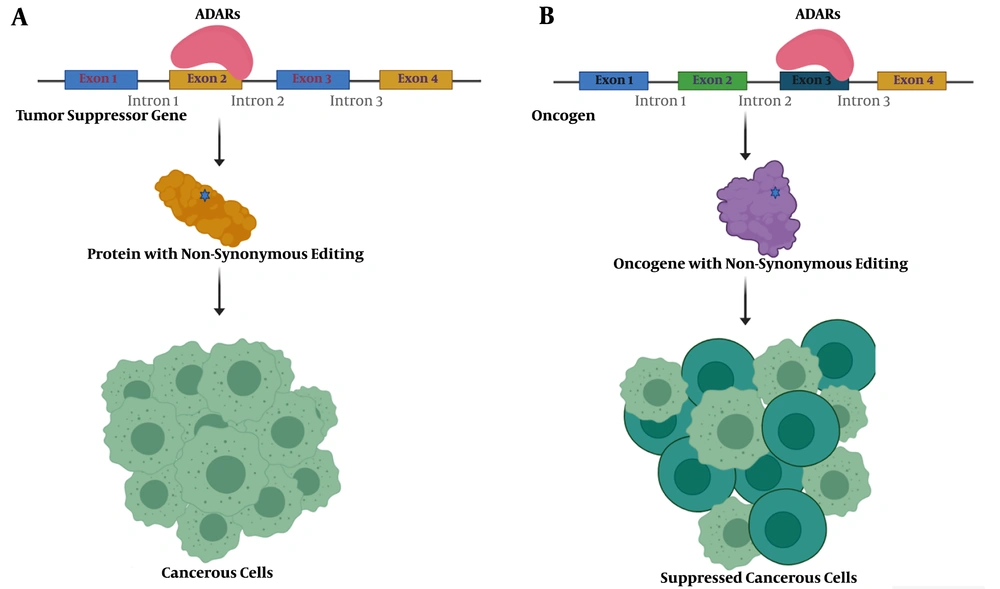
Schematic presentation of nonsynonymous editing impacts in cancer. A, a nonsynonymous editing event in the tumor suppressor gene's coding sequence could inactivate the resulting protein and lead to tumorigenesis; B, nonsynonymous editing events in activated oncogene may decrease protein activity and suppress cancerous cells.
Bladder cancer-associated protein (BLCAP), as a tumor suppressor, prevents tumorigenesis by stimulating cell apoptosis and inhibiting proliferation. ADAR1 edits several sites across BLCAP, one of them located in the nucleotide encoding the second codon with tyrosine to cysteine substitution effect (27). It has been reported that editing events in the BLCAP YXXQ motif with nonsynonymous effect drive the progression of cervical carcinogenesis through regulating the STAT3 signaling pathway (28).
3.3.2. Anti-tumorigenic Functions of RNA Editing
On the contrary, RNA editing sites may have anti-tumorigenic functions. Editing events in gamma-aminobutyric acid receptor subunit alpha-3 (GABRA3) decrease its expression on the cell surface and, consequently, prevents AKT activation required for cell migration and invasion (29). A to G editing in insulin-like growth factor-binding protein 7 (IGFBP7) at codon 95 results in amino acid recoding from lysine to arginine. This substitution arrests tumor growth and induces apoptosis in esophageal squamous cell carcinoma (30).
The Hedgehog signaling pathway is pivotal in normal cell growth and differentiation, and the glioma-associated oncogene homolog 1 (GLI1) is the central intermediary of this pathway (31). In multiple myeloma, RNA editing of GLI1 leads to alteration of arginine to glycine at 701 positions. These editing events inhibit the binding of its negative regulator (suppressor of fused homolog, SUFU) and, consequently, stabilize GLI1 expression (32). Lazzari et al. demonstrated that the malignant regenerative capacity of multiple myeloma is associated with this arginine to glycine recoding and enhances the transcriptional activity of edited GLI1 (33). Surprisingly, in medulloblastoma and basal cell carcinoma, this editing event in GLI1 acts in the opposite direction and inhibits tumorigenesis (16). These findings indicate that distinction between pro-tumorigenic and anti-tumorigenic functions of editing sites could be indeterminate even with the same recording event.
3.3.3. RNA Editing Role in Non-coding Sequences
In addition to RNA editing with the recoding effect, a significant number of RNA editing events occur in the non-coding parts of the genome (Figure 2). These sequences are involved in the post-transcriptional and post-translational regulation of gene expression, such as 3′UTRs and microRNAs (34). Sagredo et al. reported an increased number of edited sites located at 3′UTRs of ATM, GINS4, and POLH transcripts in tumors, which correlated with their mRNA expression (35). A comprehensive study across 14 cancer types suggested that changes in RNA editing levels in coding and 3′UTR regions could be a general mechanism to promote tumor growth (36). In contrast, ADAR1 edits 3′UTR of proto-oncogenes X-linked inhibitor of apoptosis (XIAP) and MDM2 and negatively regulates their protein products, serving as an anti-tumor function (37).
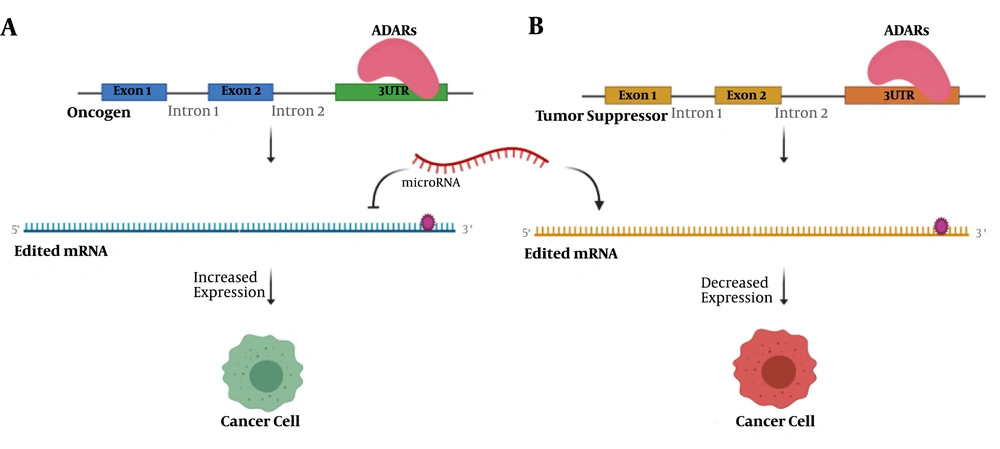
The function of 3′UTR editing in cancer initiation and progression. A, editing events in 3′UTR of an oncogene affect the binding of regulating microRNA, which in turn leads to increased expression of oncogene and tumorigenesis; B, an editing site in 3′UTR of tumor suppressor may determine it as the target of microRNA. A decreased level of this tumor suppressor could drive the cell to cancer.
A significant number of editing events in microRNAs with a functional role in cancer have been reported. Editing events on microRNAs could change their faith and function, either by preventing to regulate target mRNA or by dysregulating an off-target mRNA (Figure 3). MiR-455-5p has two different editing sites, which change the function of this microRNA. Unedited form of mir-455-5p inhibits cytoplasmic polyadenylation element-binding protein 1 (CPEB1), tumor suppressor gene, and promotes melanoma invasion and metastasis. On the other hand, the edited form of miR-455-5p represses melanoma growth and metastasis (38). Editing site on miR-589-3p changes the target of this microRNA from the protocadherin 9 (PCDH9), a tumor suppressor gene, to ADAM12, a metalloproteinase gene. The overall result is promoting glioblastoma invasion (39). Editing of miR-378a-3p occurs only in the non-metastatic but not in metastatic melanoma cells and inhibits PARVA oncogene, thus prevents the progression of melanoma towards the malignant phenotype (40). In glioblastoma cells, wild-type miR-376a promotes cell migration and invasion, while the edited form of this microRNA suppresses metastatic characteristics (41). Wild-type miR-200b regulates zinc finger E-box binding homeobox 1 and 2 (ZEB1 and ZEB2) expression and thus controls metastasis. A pan-cancer study showed that edited miR-200b could not inhibit ZEB1 and ZEB2, which results in cell invasion and migration (42).
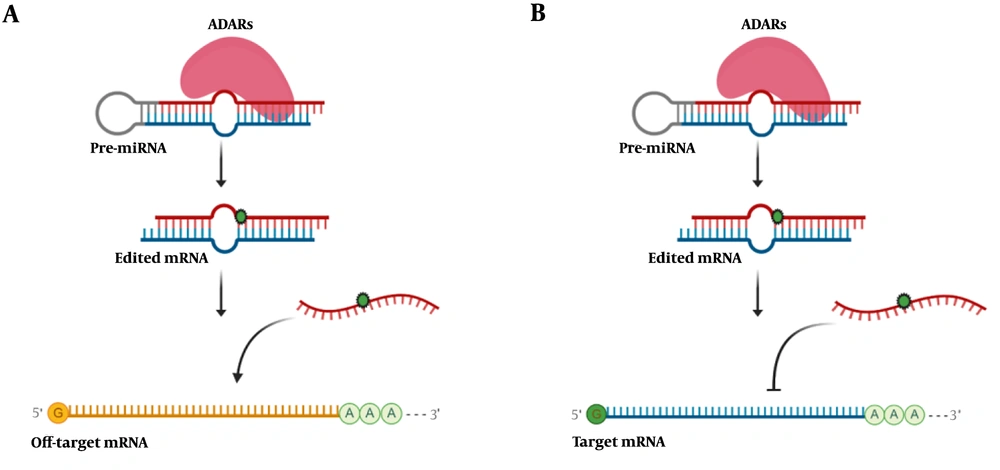
Schematic illustration of microRNA editing impact in gene expression. A, RNA editing in microRNA could change the target of that microRNA and subsequently decrease off-target mRNA expression; B, editing events interfere in microRNA base-pairing with its target mRNA and increases the expression level of target mRNA.
4. Conclusions
This study demonstrated that RNA editing is linked to cancer pathogenesis by either site specific editing of tumor-promoting/suppressing genes or by transcriptome-wide RNA editing. An increasing body of evidence indicates that RNA editing events may play an essential role in cancer initiation and progression. Editome profiling studies all pointed to a significant number of editing sites in coding and non-coding sequences. Some of these editing sites have been linked to cancer status and prognosis. Editing events with nonsynonymous effects and editing sites in 3′UTRs and microRNAs have attracted the researcher's attention.