1. Background
Nonalcoholic fatty liver disease (NAFLD) is the most prevalent liver disease in the world, associated with lifestyle and obesity. It may progress to severe hepatic disorders, including simple steatosis, nonalcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (1). Nonalcoholic fatty liver disease affects around 90% of obese patients and 25% of people worldwide (2). Nonalcoholic steatohepatitis might lead to liver cirrhosis and, subsequently, HCC. It is the second most common risk factor for HCC in patients undergoing liver transplantation in the USA (3, 4). Moreover, NAFLD is associated with other comorbidities, including metabolic syndrome, type 2 diabetes, cardiovascular disorders, and extrahepatic cancers (5).
The NAFLD pathogenesis is associated with genetic and environmental factors. Multiple tissues, including the adipose tissue, hypothalamus, intestine, and liver, are involved in the pathogenesis of NAFLD. This multifactorial disease is related to the altered bowel microbiota, increased oxidative stress (OS), and dysregulated adipokines and hormones (2, 6). Given the complexities of this disease and limited data on the underlying molecular mechanisms involved in its progression, there are no approved remedies for NAFLD treatment despite lots of efforts to detect pharmacological therapies (7, 8). Since this disorder results from lifestyle, diet, and physical activity, alterations in this regard may be helpful but not easy to achieve. Consequently, novel and effective treatments are controversial. The NAFLD diagnosis and staging are typically established by hepatic biopsy. Hence, noninvasive biomarkers that might recognize patients at risk of progression to the advanced stage are immediately necessary and helpful for drug development.
2. Objectives
Identifying gene-specific expression panels could improve our understanding of the pathogenesis and progression mechanisms of the disease or drug assessment. To the best of our knowledge, two studies have performed protein-protein interaction (PPI) networks and pathway enrichment analyses to screen differentially expressed genes (DEGs) in different stages of NAFLD compared with controls (9, 10). Wang et al. used one microarray dataset, GSE48452, to detect DEGs among control, healthy obese, steatosis, and NASH people (9). Another study used six datasets (GSE48452, GSE66676, GSE72756, GSE63067, GSE89632, and GSE107231) to find DEGs in NAFLD patients and controls. However, the GSE72756 dataset used in their study (in which normal liver tissues from NAFLD patients were used as controls) and the GSE107231 dataset do not contain three stages of NAFLD. Besides, the GSE66676 dataset contains liver tissue samples from adolescents (13.4 - 19.8 years) (10), while NAFLD in adolescents is different from that in adults (11). Thus, the present study further analyzed three microarray datasets, including GSE48452, GSE63067, and GSE89632, to explore DEGs among healthy controls, simple steatosis, and NASH patients. Furthermore, PPI networks and pathway enrichment analyses were conducted to find genes/proteins and biological pathways shared between different stages of NAFLD.
3. Methods
3.1. Data Processing
A literature search was carried out in gene expression omnibus (GEO) electronic database up to March 2021. The chosen keywords included "NAFLD," "nonalcoholic fatty liver disease," "simple steatosis," "NASH," and "nonalcoholic steatohepatitis."
Microarray studies were conducted on adults of any gender or ethnicity who met the inclusion criteria. The exclusion criteria included the studies that were randomized controlled trials, had been published before 2013, lacked a control group, reported gene expression from tissues other than the liver, and were animal, in vitro, or ex vivo research. Finally, three datasets, GSE63067, GSE89632, and GSE48452, were selected from the NCBI GEO database for the differential detection of genes (12-14). These three datasets followed the same grouping of samples.
GSE63067 was determined in samples from three groups:
(1) Nine patients diagnosed with early-stage NASH (NA),
(2) Two patients diagnosed with nonalcoholic hepatic steatosis (NA), and
(3) Seven controls (NA) with normal liver function and histology.
GSE89632 was determined in samples from three groups:
(1) Nineteen patients diagnosed with early-stage NASH (10 females and nine males with a mean age of 43.5 years and BMI of 31.9),
(2) Twenty patients diagnosed with nonalcoholic hepatic steatosis (six females and 14 males with a mean age of 44.7 years and BMI of 28.6), and
(3) Twenty-four controls (13 females and 11 males with a mean age of 37.2 years and BMI of 26.1) with normal liver function and histology.
GSE48452 was determined in samples from three groups:
(1) Eighteen patients diagnosed with early-stage NASH (14 females and four males with a mean age of 45.4 years and BMI of 45.9),
(2) Fourteen patients diagnosed with nonalcoholic hepatic steatosis (10 females and four males with a mean age of 41.6 years and BMI of 48.2), and
(3) Fourteen controls (nine females and five males with a mean age of 51.8 years and BMI of 25.14) with normal liver function and histology.
The selected datasets included microarray data processed by the LIMMA package in R, followed by performing quantile normalization and quality control and identifying and removing the noisy data and outliers by hierarchical clustering. The calculation of DEGs was done by the eBays Method. Among different data analysis methods, such as integration and fusion, the integration method was selected, involving a separate analysis of each dataset. In this method, each dataset is analyzed without any specific dependency, and finally, the results are integrated at the decision level. The DEGs with |FC| ≥ 2 and P-value ≤ 0.05 were selected as the final DEG list for all three data groups. Data were collected from three groups of healthy, steatosis, and NASH participants (Table 1).
| GSE_ID | Participants | Tissues | Platform | Year |
|---|---|---|---|---|
| GSE63067 | 9 NASH,2 Steatosis, 7 Healthy | Liver | GPL570 | 2014 |
| GSE89632 | 19 NASH, 20 Steatosis, 24 Healthy | Liver | GPL14951 | 2016 |
| GSE48452 | 18 NASH, 14 Steatosis, 14 Healthy | Liver | GPL11532 | 2013 |
For both groups in each dataset, a differential list was obtained. The lists were DEGs related to two healthy and steatosis groups named SH, healthy and NASH named NH, and steatosis and NASH named NS, respectively.
3.2. Physical Protein Interaction Network
To plot the physical interaction network, the researchers combined all groups (i.e., SH, NS, and NH) in the dataset, and then the tool https://string-db.org was used. Each list's physical and experimental interactions were examined, and the interaction network was extracted. Moreover, to study the deeper connections between gene interactions in liver tissue, the researchers entered all gene lists as input to the HIPPIE tool (http://cbdm-01.zdv.uni-mainz.de). The interaction data were extracted by filtering interactions related to liver tissue. Finally, all the extracted networks were entered into the Cytoscape (https://cytoscape.org), and Network Analyzer identified the hub genes (with the highest degree).
3.3. Similarity Analysis
In order to investigate the similarities between the three processing sets, the study selected identical groups in pairs and overlapping genes. Also, since the enrichment analysis was performed for each group, the overlaps in each analysis were examined.
3.4. Enrichment Analysis
The present study applied Enrichr (maayanlab.cloud) to find the biological pathways and the gene ontology involved in the gene list. The data were used to investigate the biological similarities of the gene sets in the gene list.
4. Results
4.1. Identification of DEGs in NAFLD
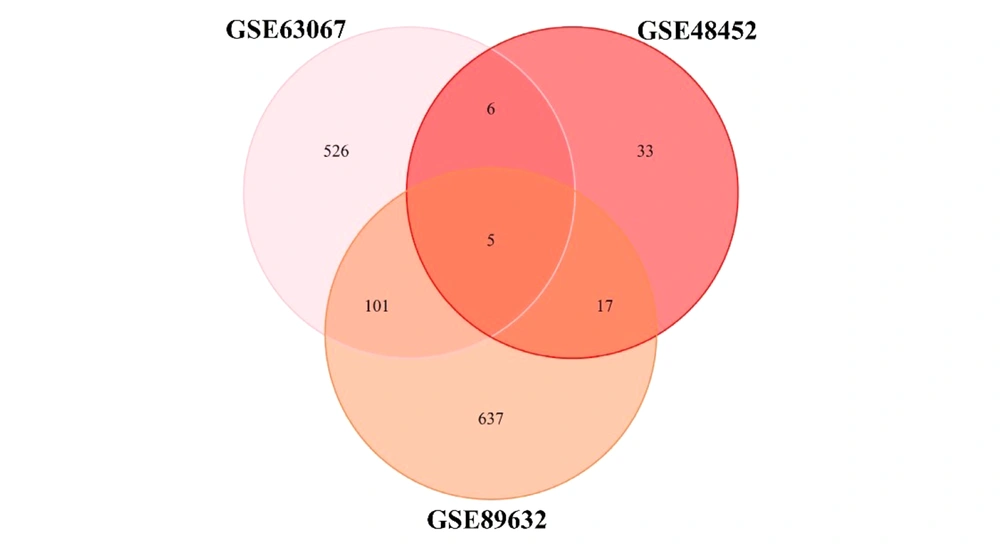
All three microarray datasets were first analyzed separately. The results of data normalization and clustering can be found in Supplementary File 1. The DEGs were screened using the "LIMMA" package in R software according to the data processing section criteria. The DEG lists of the three microarrays are shown in Supplementary Files 2, 3, and 4 for GSE48452, GSE63067, and GSE89632, respectively. The results of the three microarrays are presented in Table 2. Besides, the overlapping genes in this step are shown in Figure 1. No genes were shared between the three groups in the GSE48452 dataset (A). In the GSE63067 dataset (B), two genes were found to be shared between the three groups, Intercellular Adhesion Molecule 1 (ICAM1) and Paternally Expressed Gene-10 (PEG10). In the GSE89632 dataset (C), three genes were found to be shared between the three groups, Flavin-Containing Monooxygenases 1 (FMO1), PEG10, and Metallothionein 1A (MT1A).
| GSE_ID | GSE63067 | GSE89632 | GSE48452 | |||
|---|---|---|---|---|---|---|
| UPs | DOWNs | UPs | DOWNs | UPs | DOWNs | |
| SH | 283 | 66 | 186 | 366 | 11 | 5 |
| NS | 229 | 59 | 17 | 10 | 16 | 5 |
| NH | 120 | 19 | 244 | 321 | 24 | 10 |
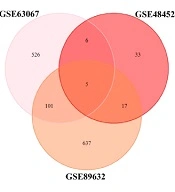
By examining three datasets to find the list of shared genes between all the three sets, the researchers came across five genes, CHI3L1 (Chitinase 3 Like 1), GFBP2 (Insulin-like growth factor (IGF) binding protein 2), NRG1 (Neuregulin 1), PEG10, and FADS2 (Fatty Acid Desaturase 2) (Figure 2).
4.2. Overlapping Genes in Matched Groups
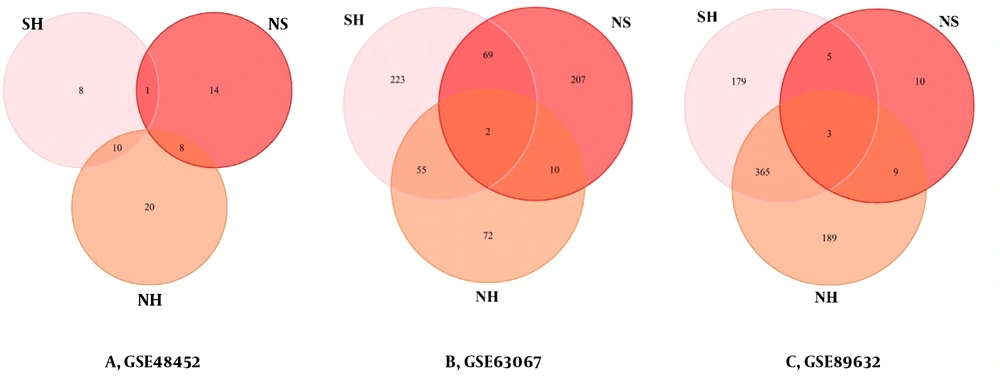
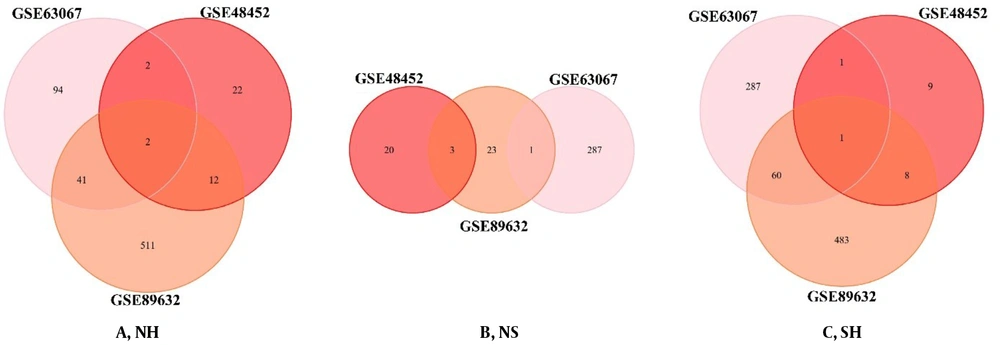
The DEGs of each processed group are used to identify the similarities between the two sets. We plotted the Venn diagram for the overlapping genes in the three groups (Figure 3). IGFBP2 (insulin-like growth factor-binding proteins) and PEG10 were the shared genes in the list of NH (Figure 3A). There were no shared genes in the list of NS (Figure 3B). There was only the PEG10 gene as the shared gene in the list of SH (Figure 3C).
4.3. PPI Networks
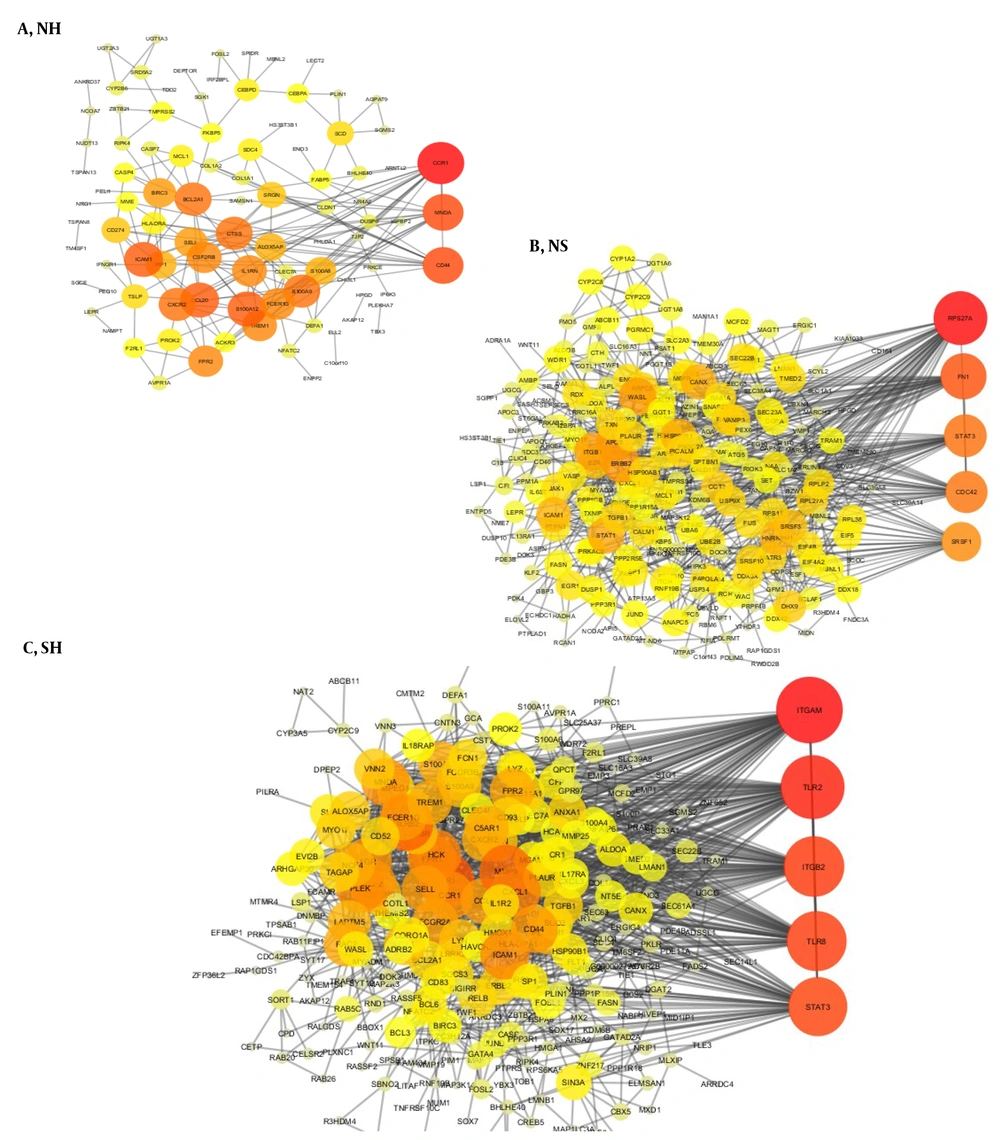
Gene interactions for each group indicated a significant number of interactions; therefore, for each group, the genes with the highest degree of interaction can be considered significant points in the network. Three global interaction networks were extracted from three gene lists that can be seen in Figures 4A - C (Red color indicates the nodes with the highest degree in this network, and as the color tends to yellow, the degree of nodes decreases).
Besides, to study the genetic connections in a more detailed way in the liver tissue, we examined these connections specifically. These liver tissue connections suggest that hub genes can play a crucial role in disease due to their location and role in the interaction network. The Signal Transducer and Activator of Transcription 3 (STAT3) and JUN genes, as the gene with the highest degree, play a crucial role in liver tissue.
Using the network analyzer plugin in the Cytoscape, we analyzed the hub genes topologically, and the top hub genes with the highest degrees were extracted using the criteria. In the interaction networks, the gene with the highest degree has the most connections and plays an intermediate role between other genes; it is known as a bottleneck. In case of any problem with the network, this intermediate gene can help understand the network's structure.
4.4. Enrichment Analysis
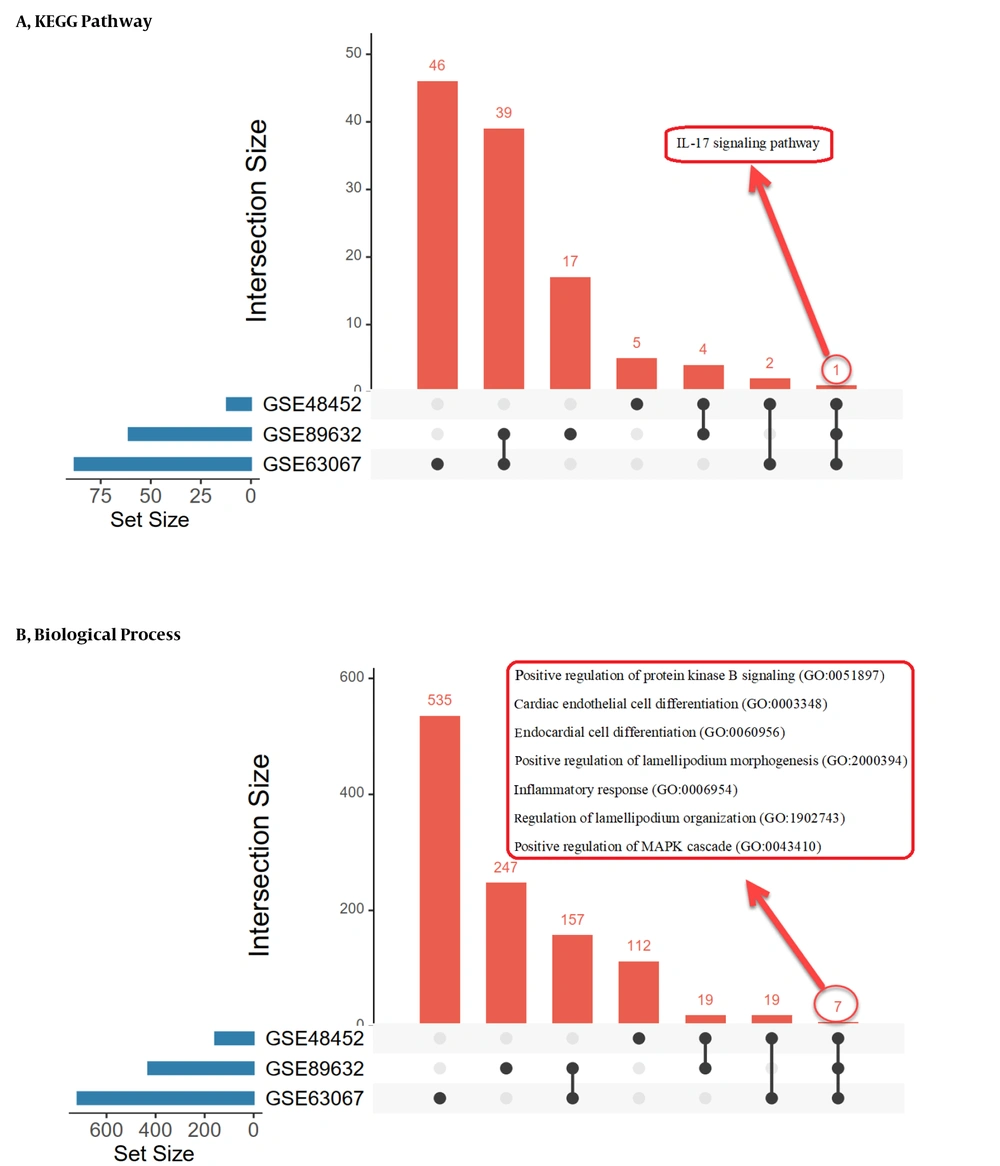
The biological pathways and processes shared between datasets are listed in Figure 5, which is the output of UpSet (https://asntech.shinyapps.io). We obtained three enrichment lists by examining the overlap between the ontologies of genes and the shared biological pathways in each group. Tables 3 - 5 show all enrichment list subscriptions.
| Biological Pathways | P-Value (Hypergeometric Test) | Bonferroni Method |
|---|---|---|
| TNF signaling pathway | 1.18E-09 | 3.10E-07 |
| Viral protein interaction with cytokine and cytokine receptor | 4.44E-08 | 3.875E-06 |
| AGE-RAGE signaling pathway in diabetic complications | 2.63E-07 | 0.00001722 |
| Cytokine-cytokine receptor interaction | 8.41E-09 | 1.102E-06 |
| IL-17 signaling pathway | 0.00001786 | 0.0007798 |
| JAK-STAT signaling pathway | 2.835E-06 | 0.0001485 |
| Malaria | 0.0002654 | 0.005349 |
| Chagas disease | 0.00004321 | 0.001258 |
| C-type lectin receptor signaling pathway | 0.00005311 | 0.001391 |
| Osteoclast differentiation | 0.00002855 | 0.001068 |
| Biological Pathways | P-Value (Hypergeometric Test) | Bonferroni Method |
|---|---|---|
| Fatty acid elongation | 0.0009678 | 0.01901 |
| Lipid and atherosclerosis | 3.302E-06 | 0.000908 |
| Proteoglycans in cancer | 8.957E-06 | 0.001232 |
| Fructose and mannose metabolism | 0.002087 | 0.02733 |
| Bacterial invasion of epithelial cells | 0.0002898 | 0.01139 |
| Leishmaniasis | 0.0002898 | 0.01139 |
| IL-17 signaling pathway | 0.0001709 | 0.01139 |
| Tight junction | 0.0001256 | 0.01139 |
| Th17 cell differentiation | 0.0004155 | 0.01217 |
| Neomycin, kanamycin, and gentamicin biosynthesis | 0.0803 | 0.2389 |
| Biological Pathways | P-Value (Hypergeometric Test) | Bonferroni Method |
|---|---|---|
| IL-17 signaling pathway | 4.33E-11 | 1.18E-08 |
| Leishmaniasis | 2.58E-09 | 1.17E-07 |
| TNF signaling pathway | 2.64E-10 | 3.58E-08 |
| Osteoclast differentiation | 6.54E-10 | 4.45E-08 |
| Malaria | 8.67E-07 | 0.00002621 |
| Rheumatoid arthritis | 6.07E-08 | 0.000002359 |
| Transcriptional misregulation in cancer | 6.43E-10 | 4.45E-08 |
| Viral protein interaction with cytokine and cytokine receptor | 1.93E-07 | 0.000006571 |
| Legionellosis | 0.000003845 | 0.0000747 |
| Inflammatory bowel disease | 0.000002811 | 0.00005882 |
The most important terms of gene ontology in the biological process are signal transduction, immune response, and anti-apoptosis in the NH group, metabolism and energy pathways in the NS group, and cell communication, signal transduction, and immune response in the SH group. They are also parts of the pathways that significantly overlap in the study groups.
Although the results obtained from data fusion are consistent with individual data, it is noteworthy that reviewing the results in different datasets can be very helpful in confirming the results.
5. Discussion
In the present study, PPI networks were produced based on differential gene expression provided from liver tissue of simple steatosis and NASH patients compared with healthy controls. Centrality and modularity analyses of PPI networks helped us detect common genes and pathways involved in these diseases. In order to evaluate the results based on the at-hand evidence, we used the Human Protein Atlas Database, and the results were matched with the list of approved proteins. The results revealed that many proteins had been examined in the experiments.
Our findings suggested three overlapping genes in three groups (Chi3L1, ICAM1, and MT1A), whereas two groups shared several genes. A biological pathway was also shared by all groups (IL-7). Herein, these genes' roles in various disease phases are explained. Intercellular Adhesion Molecule 1 (ICAM1) is a glycoprotein located on the cell surface and expressed in some cells such as liver, endothelial, epithelial, and hematopoietic cells. The overexpression of ICAM1 may occur by some inflammatory cytokines, including IL-1 and TNF-α (15). Ito et al. suggested that serum ICAM1 might be a diagnostic marker for NASH as its concentration increases in NASH patients compared with healthy individuals, which aligns with our finding. Moreover, there was a positive correlation between serum ICAM1 levels and the severity of the liver injury and inflammation (15). Another study showed that hepatic expression of ICAM1 is significantly higher in NASH patients than in simple steatosis and normal controls. Moreover, a significant correlation was observed between the steatosis degree and hepatic ICAM1 expression (16). It has been hypothesized that overexpression of hepatic ICAM1 enhances leukocyte adherence and increases Reactive Oxygen Species (ROS) production, indicating the severity of necroinflammatory activity (17).
The molecular processes linked with steatosis development and disease progression were investigated in this study. Our findings presented the most critical genes in the liver tissue associated with the progression of different stages of NAFLD. Bile acids may play a role in the development and treatment of NAFLD/NASH by signaling through their designated nuclear receptor (i.e., farnesoid X receptor) (FXR; NR1H4) as a critical regulator of glucose and lipid metabolism, as well as inflammation (18, 19). The bile salt export pump (BSEP and ABCB11) excretes bile acids from hepatocytes into the gallbladder. In humans, there has been evidence of links between BSEP polymorphisms and elevated serum triglycerides (TG), cholesterol, and obesity (20). When mice overexpressing BSEP are fed a lithogenic diet or a methionine choline-deficient (MCD) diet, they develop moderate hepatic steatosis (21). Diminished expression of ATP Binding Cassette Subfamily B Member 11 (ABCB11), and therefore reduced BSEP pump, were also found to be one of the causes of steatosis progression to NASH, according to the findings of Okushin et al. (22). In fact, lowering the expression of this pump causes an increase in bile acids in liver cells, resulting in cell damage (22). Our findings also revealed that the expression of the ABCB11 gene in simple steatosis was decreased compared to healthy persons, suggesting that this drop in expression could be one of the crucial elements in the development and progression of fatty liver disease. These findings were also confirmed in transthyretin-ABCB11 (TTR-ABCB11) mice, which have much greater levels of ABCB11 compared to the wild ones. The findings of investigations suggest that this type of mouse develops hepatic steatosis less than a regular one (23, 24).
At complexes I and III of the electron transport chain, mitochondrial Fatty Acid (FA) oxidation creates a small amount of ROS (25). Antioxidant activity counteracts this ROS generation, protecting the mitochondria from OS (26). On the other hand, high membrane potential might cause a substantial amount of ROS by increasing FA oxidation, enhancing antioxidant capacity, and resulting in oxidative stress (27). Controlling the rate at which FAs are oxidized in a cell is essential for reducing ROS and oxidative stress that might arise with an increase in oxidation. Acyl-CoA thioesterase 1 (ACOT1) catalyzes the conversion of acyl-CoA to FAs and CoA, which subsequently limits OS (28). In mice with diabetic cardiomyopathy, overexpression of ACOT1 in cardiomyocytes decreases FA oxidation and ROS generation (29). These findings imply that ACOT1 controls FAs destined for oxidative pathways, presumably by eliminating acyl-CoAs as substrates. Also, ACOT1 may protect the liver from the harmful consequences of excessive FA oxidation by delaying FA oxidation (28). Moreover, PPARα has been shown to have anti-inflammatory and protective effects in inflammatory disorders of the liver, and ACOT1 leads to the supply of ligands required for its activation. Consequently, enhanced ACOT1 expression may help prevent steatosis and the development of steatosis to other inflammatory liver disorders (28, 30). Our study also indicated that the expression of the ACOT1 gene is lower in steatosis than in healthy conditions, suggesting that this decrease in expression might be one of the causes of FA metabolism disturbance and the beginning of the fatty liver disease.
Chitinase 3-like 1 (Chi3L1), a newly identified human glycoprotein, has a role in inflammation, tissue remodeling, and visceral fat storage (31). Increased Chi3L1 gene expression is a factor in increasing insulin resistance and, thus, fat accumulation in the liver. In fact, mice with the Chi3L1 gene knockout are more insulin sensitive, which slows the progression of the disease to NASH (32). Chi3L1 is generated by various inflammatory cells and can enhance the production of inflammatory cytokines such as IL-13, IL-6, IL1, and IFN-γ (33). Chi3L1 serum levels have been linked to different phases of liver fibrosis in recent clinical trials (34, 35). According to preliminary research, Chi3L1 gene expression can discriminate between isolated simple steatosis and NASH by whole genome RNA sequencing compared to existing biomarker scoring systems (RNA-seq) (32). Our results also showed that the expression of the Chi3L1 gene is higher in patients in the NASH stage of fatty liver disease than in simple steatosis. Therefore, this gene can be used as a biomarker to detect the progression of NAFLD.
As known, CYP1, CYP2, and CYP3 are members of the cytochrome P450 (P450) enzyme family that are responsible for the metabolism of roughly 75% of all therapeutically relevant medications. With the rising incidence of NAFLD, individuals with this condition are expected to be a new group at risk for changes in these critical drug-metabolizing enzymes (36). One of the CYPs important in the metabolism of endogenous and exogenous substances is CYP2C9. Fisher et al. (36) reported that hepatic mRNA and enzymatic activity of CYP2C9 increased with progressive stages of NAFLD, per our findings. CYP2C9 activity increases in hypoxic conditions (37). Studies have shown that hypoxia is associated with NAFLD severity and provides a plausible reason for the enhanced CYP2C9 activity in human and animal models (36, 38). Hypoxia can also cause inflammation, a feature of NAFLD (39).
Sterol regulatory element-binding proteins (SREBPs) are critical regulators of lipogenesis, regulating the expression of genes involved in fatty acid production and uptake, as well as cholesterol and phospholipid uptake (40). Heat shock protein 90 (HSP90) is a protein family that has a role in various pathophysiological events. These proteins, particularly HSP90B1, are also involved in lipid homeostasis via the SREBP pathway (41, 42). Previous studies indicated that HSP90 gene expression increases in the inflammatory stages of NAFLD disease, and inhibition of this protein by factors such as 17-AAG could reduce disease progression (43). These findings were also confirmed in our research, suggesting that in the NASH stage of the disease, the expression of HSP90 family proteins, particularly HSP90AB1 and HSP90B1, increases compared to simple steatosis.
Another hub gene, MT1A, is a family of Metallothioneins with a high affinity for binding essential and toxic metals such as zinc and cadmium, respectively (44, 45). The biological functions of MT1A are protection against toxic metals, defense against OS, and protection against cytotoxicity and genotoxicity (45-47). Additionally, MT1A has an indispensable role in the regeneration of hepatocytes (48). In line with our results, Arendt et al. demonstrated the down-regulation of MT1A gene expression in patients with simple steatosis compared to healthy controls; they observed a further decrease in NASH patients compared to patients with simple steatosis (13).
Carboxypeptidase B2 (CPB2) is another protein discovered in both databases. The liver produces plasma carboxypeptidase, which is transformed into the active enzyme by thrombin (49). To the best of our knowledge, there is not enough data on its link to human liver diseases. As a result, further research on this protein in different stages of liver illnesses is recommended.
One biological pathway, IL-7, was shared by all groups, as tabulated in Tables 3-5. T-cell development has been demonstrated to be dependent on IL-7 (50). It has been suggested that IL-7 and its receptor (CD127) have a role in NAFLD development. In NAFLD, serum IL-7 levels were inversely linked with fibrosis (51). In addition, the liver is a crucial producer of IL-7 in response to TLR stimulation. The TLR signaling in Kupffer cells drives the transition of simple steatosis to NASH (52). The gut microbiome can produce TLR ligands. TLR ligands can induce the production of proinflammatory cytokines in liver cells. Using probiotics or prebiotics to normalize the gut flora is a viable therapy option for NAFLD (53).
The strength of our study is that our findings are based on a combination of three datasets, and the genes discussed are those found to be shared in all three datasets. However, this study also has some limitations, including lacking an experimental section. For future studies, we recommend that the researchers carefully evaluate the expression of the proposed genes and proteins involved in developing NAFLD by experimental methods such as qRT-PCR.
This study revealed unique vital genes found in both simple steatosis and NASH. After further verification, the shared genes (Chi3L1, ICAM1, MT1A, MT1H, ABCB11, ACOT1, CYP2C9, HSP90B1, and CPB2) could be used as biomarkers of NAFLD progression and a putative candidate for therapeutic targets in NAFLD. Moreover, pathway analysis found that DEGs significantly enriched in several pathways are involved in inflammation and oxidative stress pathways.