1. Context
Metastasis is the leading cause of death among cancer patients that is responsible for up to 90% of cancer deaths (1). Basically, Overall survival (OS) of patients with early stages is higher than the advanced stages that metastatic sites are a vital factor on the OS. In advanced breast cancer, the 3-year survival rate of patients with bone metastasis is 50.5%, whereas in patients with brain metastasis, it is 27.7% (2). Metastasis is a complex process that enables primary tumor cells to spread systemically and develop secondary tumor mass in different sites of the body (3). This cascade process consists of local invasion, intravasation, circulation, extravasation, micrometastasis, and macrometastasis. However, cancer cells for local invasion have two types of migrations, namely collective and single-cell migration. In collective migration, a cluster of tumor masses that bind with each other migrate to sites near the tumor mass. Single-cell migration occurs through amoeboid and mesenchymal (or EMT) movement. Cells leave tumor mass and migrate local sites solitary in both migration types (4).
In such a way, derived factors of the microenvironment push tumor cells to induce EMT (5). In addition, TME-derived factors and their effects on EMT have an insightful part in the literature. Intrinsic and extrinsic factors that have affected EMT were the focus of D’Angelo et al. (6). Likewise, Erin et al. (7) stress drug resistances and EMT that are modulated by TME-derived factors. Besides, gene expression (8) and epigenetic modification (9) of EMT are other subjects for reviewing in the articles. Some review articles considered EMT mechanisms (10) involved-signaling pathways (11) and metabolic pathways that take part in EMT (12). Recent research has focused on TME cells and their derived factors that induce EMT through signaling pathways to manifest different steps of a path that is started with TME cells and finished with EMT. Herein, we firstly clarify the EMT process and the molecular sequence that changes the behavior of the tumor cells. Microenvironment cells and their products that induce EMT based on signaling pathways are discussed in the following section. The last section discusses the inducers and signaling pathways that are involved in EMT.
2. Evidence Acquisition
National Center for Biotechnology Information (NCBI) was the main resource in the current study. The review was conducted, using keywords such as ‘metastasis’, ‘invasion’, ‘epithelial-mesenchymal transition’, ‘EMT’, ‘tumor microenvironment’, ‘TME’, ‘TME cells’, and ‘signaling pathway in EMT’ thorough search in PubMed, ScienceDirect. Google Scholar and Scopus were other databases for finding articles. Titles and abstracts of the articles were studied to choose the appropriate articles. Furthermore, the latest articles and books were reviewed.
3. Results
3.1. EMT Process
Elizabeth Hay was the first researcher, who described the conversion of epithelial cells to mesenchymal cells during development and termed the process “epithelial-mesenchymal transformation” and the reverse process as “mesenchymal-epithelial transformation” (13). Greenburg and Hay (14) proposed that epithelial to mesenchymal transformation occurred in embryonic and adult epithelial cells. Meanwhile, the term ‘transformation’ was changed to ‘transition’ to distinguish from neoplastic transformation (15). Epithelial-mesenchymal transition (EMT) is a biological process, during which epithelial cells attenuate their epithelial features and acquire mesenchymal features. EMT appears in development, wound healing, and metastasis. During EMT, the cytoskeleton architecture is remodeled; so, cells lose their apical-basal polarity and cell-cell adhesion, which are primarily epithelial features. Then, they become individualized and gain mesenchymal features such as motility and invasion (16). Apart from that, tumor cells during EMT have not completely shifted from epithelial state to mesenchymal state. Indeed, EMT is an elastic and continuum process that enables cells to synchronously co-express epithelial and mesenchymal features. Therefore, EMT refers to a spectrum of epithelial and mesenchymal phenotypes that correspond to intermediate or partial EMTs (10). EMT can be classified into 3 main types depending on the process they are involved. Type 1 occurs in embryogenesis and organ development, in which epithelial cells that underwent EMT have the potential to earn their epithelial features and generate mesenchymal-epithelial transition (MET). Secondary, type 2 seems in wound healing, tissue regeneration, and organ fibrosis that inflammatory signals are a trigger for starting this EMT. Finally, type 3 of EMT transpires in cancer progression and metastasis that creates neoplastic cells to migrate and invade in closed and distant tissues (17, 18). Broadly, the critical steps of the EMT process are loss of tight cell-cell adhesion complex, changing cell polarity, and enabling migration and invasion (19). Initially, cells that undergoing EMT rupture their interaction with the basement membrane (BM) and in the next step lose both cell-cell adhesions and epithelial sheet integrity. In the following, cells rearrangement cell-BM interactions and reorganize different metabolisms that are coped with mesenchymal phenotypes. Whence, epithelial cells that were interconnected to other cells with a columnar shape lose their cell-cell adhesion and earn a spindle-like shape that can travel through extracellular matrix (ECM) (20).
3.2. Genes, Epigenetics, and Proteins Switches in EMT
During EMT, multiple cellular mechanisms change in different phases of gene expressions, epigenetic, and pathways (Table 1) that lead tumor cells to earn new features that promote tumor progression and metastasis (10). At the epigenetic level, the miR-200 family inhibits EMT through the degradation of mRNA factors that are promoted EMT. However, expression of miR-200 family down-regulates in different cancers that is a proper condition for up-regulation of mesenchymal markers (12). Different microRNAs like miR-9 and miR-10b down-regulate epithelial markers and lead motility and metastasis in tumor cells (12). At the protein and gene levels, the expression of genes that encode epithelial and mesenchymal proteins changed during EMT (21). Gene expression of epithelial markers, such as E-cadherin, ZO1, laminin1, occludins, desmoplakin, and cytokeratins are down-regulated, but the expression of mesenchymal markers like N-cadherin, β-catenin, vimentin, and fibronectin are up-regulated (22). BMI-1, a protein that silences genes through the regulation of chromatin structure (23), up-regulates in tumor cells (24). BMI-1 up-regulation leads to the promotion of invasion through down-regulating of E-cadherin in nasopharyngeal carcinomas cells based on the suppression of PTEN expression and activation of PI3K/AKT/Snail signaling pathway that is forwarded cells to EMT (25).
| Markers | Up-regulation | Down-regulation | References |
|---|---|---|---|
| Adhesion-Involved markers | N-cadherin, α5 integrin, vimentin, αSMA | E-cadherin, ZO-1, cytokeratin, occludin, desmoplakin, crumbs3, PALS1, PATJ, Plakophilin | (8, 12, 22, 26) |
| ECM proteins | Collagen, fibronectin, laminin 5 | Laminin 1 | |
| Transcription Factors | Snail, Slug, ZEB1, ZEB2, Twist1, FOXC2, FOXD3, SOX9 | Grhl2, Ovol1/2 | |
| MicroRNA | miR-9, miR-10b, miR 155, miR 491 5p, miR-661 | miR-1, miR-29b, miR-30a, miR-34, miR-200 family |
3.3. Involved Transcription Factors in EMT
The orchestra changing in EMT mediates by transcription factors (TFs) (Table 1) that suppress epithelial markers and induce mesenchymal markers (8). Three master TFs, SNAIL, TWIST, and ZEB, with other TFs such, FOX, SOX, PRX, and HMGA2 lead cells forward consensus differentiations that make tumor cells capable of motility and metastasis (8). SNAIL1 (known as a snail) and SNAIL2 (known as a slug) activation down-regulates the expression of genes involved in tight junctions (occludin and claudin 1), apical polarity (CRB3), and other cell adhesions like CDH1 (encodes E-cadherin), but up-regulates the expression of N-cadherin, fibronectin, vimentin, and MMPs through different signaling pathways (27). Thereby, cellular changes that modulate by ZEB1 (known as δEF1) and ZEB2 (known as SIP1) are like snail and slug (27). Expression of E-cadherin, β-catenin, and γ-catenin mask by TWIST1, but the expression of vimentin is increment (28).
3.4. Tumor Microenvironment
Tumor mass consists of heterogeneous tumor cells and a variety of non-tumoral cells that provide a condition for the growth and progression of tumor mass (29). The composition of non-tumoral cells and secreted factors with the ECM is known as tumor microenvironment (TME) (30) that is a dynamic environment with important roles in different stages of cancer evolution (31). TME cells and derived factors have a variety among cancer types that lead processes in tumor cells to cope with new sites and construct tumor mass (31). Chiefly, the cellular residue of TME consists of stromal cells that include endothelial cells, pericytes, fibroblasts (32), and immune cells (33). Secreted factors of TME include cytokines, chemokines, and growth factors that represent network interactions between tumor cells and non-tumoral cells in TME (34). Besides, derived factors and induced signals of the TME are involved in different aspects of cancer such as tumor growth, progression, EMT, metastasis, and multidrug resistance (7). Additionally, factors that are derived from TME cells activate signaling pathways to begin the cascades for depriving epithelial markers and attenuating mesenchymal markers for drug resistance and metastasis in epithelial cells (7).
3.5. TME Cells
Microenvironment cells secret factors (Table 2) that induce EMT and metastasis in tumor cells (35). TME cells emit factors to trigger signaling pathways that activate TFs for inducing EMT, migration, and invasion (36). Infiltrating immune cells are a critical part of TME that consist of tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), dendritic cells (DCs), natural killer (NK) cells, mast cells, granulocytes, and different types of lymphocytes (37). Macrophages are one of the stromal immune cells that induce EMT and invasion in tumor cells (38). Likewise, TAMs have higher accumulations in metastatic colorectal cancer than non-metastatic colorectal cancer that their depletion decrease metastatic behavior of colorectal cancer cells to the lung. Besides, TAMs provoke EMT in colorectal tumor cells through increasing phosphorylation of SMAD2/3 that are mediated by TGF-β (39). Although, TAMs through the secretion of cytokines like IL-6, induce EMT in intrahepatic cholangiocarcinoma (ICC). The secreted ILs-6 inhibits E-cadherin and ZO-1 and augment vimentin, α-SMA, and N-cadherin based on the activation of the Akt pathway (40). Furthermore, macrophage-fused cells with breast cancer cells suppress apoptosis and promote proliferation and invasion. Also, the fused cells evoke mesenchymal markers and restrain epithelial markers by stimulation of the Wnt/β-catenin pathway (41). Otherwise, neutrophils are part of immune cells in TME that mediate EMT by secretion of inflammatory factors. Neutrophils that are stimulated by gastric cancer cells secrete inflammatory factors IL-6, 8, and TNF-α to elicit vimentin, slug, and subduing E-cadherin. As well, neutrophils that present in TME of gastric cancers lead EMT through phosphorylation of ERK. Yet, ERK inhibitor, U0126, reverse induced EMT by neutrophils that are shown ERK pathways lead EMT in gastric cells (42). Furthermore, neutrophils induce EMT in oral squamous cell carcinoma through TGFβ and IL-17A (43). On the other hand, mast cells are involved in EMT. The interaction between mast cells and non-small cell lung cancer (NSCLC) cells activates mast cells that induce EMT by IL-8 to trigger the Wnt/β-catenin signaling pathway (44). Besides, mast cell-derived extracellular vesicles (EVs) augment the expression of TWIST1, VIM, and SMAD2, which lead to EMT in lung cancer cells (45). Into the bargain, activated mast cells secrete IL-8 to trigger Akt signaling pathway that up-regulated slug expression for inducing EMT and stemness in thyroid cancer cells (46). One of the TME cells is fibroblasts that is present in connective tissues and involves ECM deposition, epithelial differentiation, and inflammation. Fibroblasts within tumor stroma modify and became activated, which are called carcinoma-associated fibroblasts (CAFs) (47). CAFs activate twist1 to induce EMT mediated by STAT3 and ERK1/2 signaling pathways in gastric cancer cell lines (48). Besides, CAFs in TME of gastric cancer cells increase the invasion of tumor cells by IL-6. CAFs-derived IL-6 promotes EMT-dependent metastasis based on the activation of the JAK2-STAT3 pathway. Phosphorylation of JAK2 and STAT3 activates ZEB2, which causes suppression of E-cadherin and augmentation of N-cadherin (49). In bladder cancer, IL-6 that emits by CAFs down-regulates epithelial markers and up-regulates mesenchymal markers; so, invasion and migration of bladder cancer cells increase through co-culturing with CAFs (50). Besides, expression level of markers like as N-cadherin, vimentin, fibronectin, and a-SMA are increased in hepatocellular carcinoma (HCC) after CAF treatment. CAFs through IL-6 activated STAT3 to induce EMT in HCC cells (51). Furthermore, CAFs induces EMT in breast cancer cells (52). CAFs increase metastasis of endometrial cancer cells and induce EMT by secretion of EGF, TGF-β, HGF, and FGF-2 (53). In lung cancers, CAFs down-regulate miR-33b expression levels and elevate the expression of snail1, twist1, and ZEB1 to induce EMT (54). CAFs activate JAK2/STAT3 and hedgehog signaling pathways in lung cancer cells to induce EMT through IL-6 (55) and direct interaction (56), respectively. Moreover, TGF-β that is generated by CAFs triggers TGF-β signaling pathway to restrain epithelial features and to augment mesenchymal features in NSCLC (57). Platelets promote EMT in colon cancer and breast cancer cell lines MC38GFP and Ep5, respectively, by secretion TGF-β1 in vitro. Also, Pf4-cre+; TGFβ1fl/+ mice that are TGFβ1-deficient platelets show lower metastasis in compared wild-type mice. Besides, platelet-derived TGF-β that promotes EMT through TGFβ Signaling synergize up-regulation of mesenchymal markers and down-regulation of epithelial markers based on the activation of NF-κB signaling pathway (58). Otherwise, endothelial cells emit HGF into TME of breast cancer that evokes vimentin and N-cadherin and restrains E-cadherin in basal-like breast cancers (59).
Abbreviations: CAFs, cancer-associated fibroblasts; TAMs, tumor-associated macrophages.
3.6. Secreted Factors of TME Cells with Their Pathways to Lead EMT
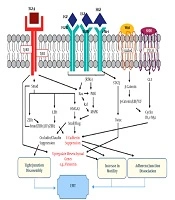
The cascade of events that happened during EMT depends on inducers and signaling pathways (Figure 1) that are secreted by TME cells and activate snail, slug, ZEB1, ZEB2, TWIST to augment mesenchymal markers and restrain epithelial markers (61).
Secreted factors and signaling pathways are involved EMT [modified (61)]
3.7. Transforming Growth Factor Beta
Transforming growth factor beta (TGF-β) is one of the growth factors that induces EMT (62) and affects tumor growth, cell survival, and tumor invasion (63). TGF-β through different pathways, such as MAPK, PIK3C, and SMAD down-regulates epithelial features and up-regulates mesenchymal features (64). The TGF-β/SMAD pathway is one of the known pathways that leads to EMT in different cancer types. According to the SMAD pathway, TGF-β binds to its heterodimeric type I and type II receptors to trigger activation of type II receptors that phosphorylate seine/threonine rich site (GS region) of type I. Following, activated type I phosphorylates serine residue in the C-terminal domain of R-SMAD (Smad2, 3) proteins. During the next step, R-SMAD constructs a complex with SMAD4 and translocates it into the nucleus. In the last step, the complex of R-SMAD and SMAD4 induces expression of snail, slug, and ZEB1/2 that suppress epithelial gene expressions (E-cadherin, ZO1) and promote mesenchymal gene expressions (vimentin, fibronectin) (63).
TGF-β through phosphorylation of Smad2 subdue EMT in breast cancer cells (65). Inhibition of TGF-β1 by tranilast that suppressed phosphorylation of Smad4 diminished mesenchymal marker and augment epithelial marker in lung cancer cells (66).
3.8. Epidermal Growth Factor
Epidermal growth factor (EGF) invokes EMT (67), tumor growth, cell survival, and cell motility in different cancer types (68). When EGFs bind to its homo or heterodimer tyrosine kinase receptors (EGFR), residues on receptors through trans-autophosphorylation became phosphorylated and adaptor proteins (Grb2, SHC) attach to the phosphorylated receptors (69) to activate signaling pathways such as PIK3C/Akt, MAPK/Erk, and STAT3 (70). In the STAT3 pathway, signal transducer and activators of transcription (STAT) proteins bind to phosphorylated tyrosine residues on receptors and became phosphorylated. Phosphorylated STAT3s dimerize and translocate into the nucleus (71) to induce the expression of genes such as twist, ZEB1, and snail (72). Thence, N-cadherin and vimentin up-regulate and E-cadherin down-regulate by STAT3 activation (73). In addition, EGFs increase the expression of Snail and mesenchymal markers through the phosphorylation of Smad2/3 in breast cancers (74).
3.9. Fibroblast Growth Factors
Fibroblast growth factors (FGFs), the same as TGF-β and EGF, induce EMT through sort of signaling pathways. FGFs as well as other EGF member families, hepatocyte growth factor (HGF), and platelet-derived growth factor (PDGF) lead tumor cells toward EMT through PIK3C, Ras/Raf/MAPK, and Src/STAT pathways (75). Ras/Raf/MAPK is one of the important pathways to induce EMT that is activated by FGF. Firstly, FGFs bind to its tyrosine kinase receptor and induce receptor dimerization. Next, trans-autophosphorylation happens and some tyrosine residues became phosphorylated on the receptors (76). In the third step, activated receptors phosphorylate FRS2α that is an anchorage for adaptor protein, Grb2, and recruiting other proteins, SOS, that remove GDP from Ras proteins (77). In the next step, Ras activates through binding of GTP. Following, Ras activates a kinase called Raf that phosphorylates serine residue of a mitogen-activated protein kinase (MAPK) known as MEK (MAPK/ERK kinase), which is a serine/threonine kinase. In the final step, activated MAPK phosphorylates tyrosine and threonine residues on the regulatory site of another MAPK called extracellular signal-regulated kinase1/2 (ERK 1/2) that translocate into the nucleus and activates transcriptions factors to induce expression of genes involved in EMT (78, 79). So, mRNAs and proteins of vimentin, α-smooth muscle actin (SMA), N-cadherin, and fibroblast-specific protein 1 (FSP1) increase in different tumor cells after FGFs treatment. On the other hand, FGFs decreased the expression of epithelial markers E-cadherin and cytokeratin (80, 81). The combination of FGF and TGF-β1 or FGF alone leads lung cancer cells toward EMT (82).
3.10. Hepatocyte Growth Factor
Hepatocyte growth factor (HGF) is one of the extracellular factors that is used in different signaling pathways to induce EMT. HGF invokes EMT according to activate STAT3 (83), Ras/MAPK (84), and PIK3C/Akt signaling pathways (85). Like other tyrosine kinase receptors, trans-autophosphorylation befall upon HGF bind to its receptor (c-MET). Next, phosphorylated c-MET became a dock for the SH domain of phosphatidylinositol 3 kinase (PI3K), which is a lipase kinase. In addition, PI3K activation intercedes with adaptor proteins and Ras. In PIK3C/Akt pathway, activated PI3K proteins phosphorylate phosphatidylinositol 4,5-bisphosphate (PIP2) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). Then, PIP3 mediates the first step activation of Akt, a serine/threonine kinase, via 3-phosphoinositide-dependent protein kinase-1 (PDK1) that phosphorylates threonine residues on Akt. Full activation of Akt gain through mammalian target of rapamycin complex-2 (mTORC2) that phosphorylates serine residues on Akt. Then, full activated Akt phosphorylates effector proteins mTORC1, TSC1/2, and RHEB are triggered for activation of cascades that are involved in cell survival, growth, EMT, and metastasis (83, 85, 86). In this manner, HGF based on snail and slug up-regulation leads to cellular changing to suppress E-cadherin and promote N-cadherin, fibronectin, and vimentin. In that, motility and invasion of tumor cells are promoted in response to HGF secretion (87). Suppression of HGF decreases invasion and migration of oral squamous cell carcinoma (OSCC). Curcumin abolishes phospho-c-Met level that caused down-regulation of HGF signaling. Therefore, curcumin through HGF signaling inhibition suppresses EMT (88).
3.11. Platelet-derived Growth Factor
Platelet-derived growth factor (PDGF) is one of the growth factors that leads to tumorigenesis. Besides, PDGF invokes EMT in colorectal cancer cells that suppression of E-cadherin and promotion of vimentin spawn Notch1/Twist1 pathway (89). Loss of E-cadherin and ZO-1 and induction of vimentin are a result of PDGF overexpression in prostate cancer cells. PDGF leads prostate cancer cells toward EMT through NF-κB and mTOR pathways that each pathway targets different genes that are involved in EMT and invasion (90).
3.12. Insulin-like Growth Factor
Insulin-like growth factor (IGF) like other mentioned growth factors yields EMT. IGF-1 mediates EMT in multiple myeloma based on PI3K/Akt signaling pathway (91). Besides, Akt/ERK pathways activated by IGF down-regulate E-cadherin and up-regulate vimentin and ZEB1 in gastric cancer cells (92).
3.13. Inflammatory Cytokine Interleukin-6
Inflammatory cytokine interleukin-6 (IL-6) is associated with EMT. Down-regulation of epithelial markers and up-regulation of mesenchymal markers by IL-6 is documented in prostate cancers (93), colon cancers (94), thyroid cancers (95), and biliary cancers (96). IL-6 induces vimentin and restrain E-cadherin through STAT3 activation in cervical and breast cancers (97, 98). In ovarian cancers, IL-6 through JAK2/STAT3 induces EMT. In JAK2/STAT3 pathways, the binding of IL-6 to its receptors activates Janus kinase-2 (JAK2) that are attached to the receptor. After that, tyrosine residues on receptor phosphorylate by JAK2. Then, phosphorylated receptors recruit STAT3 proteins through their SH2 domains. Next, JAK phosphorylates STATs translocate into the nucleus after dimerization to induce EMT (99). Furthermore, other inflammatory factors, tumor necrosis factor-alpha (TNF-α), IL-1, and IL-8, implicate on stimulation of mesenchymal markers and deprive epithelial markers in cancer types (100).
3.14. Wnt
Wnt/β-catenin signaling is an EMT signaling pathway that is activated by intercellular factors. After Wnt binding to its receptors, Frizzled (FZD), some proteins became activated that lead to the accumulation of β-catenin in the cytoplasm. Then, elevated β-catenin leaves cytoplasm and translocate into the nucleus to form a complex for activation of targeted genes such as twist and slug (11). In breast cancer, Wnt signaling inhibits glycogen synthase kinase 3β (GSK3β) to increase the level of slug in the nucleus. Up-regulation of slug decreases expression of E-cadherin and increases expression of vimentin (101). Furthermore, inhibition of Wnt/β-catenin signaling in ovarian cancer cells attenuates the mRNA level of vimentin and augment E-cadherin. In addition, migration and proliferation of ovarian cancer cells are decreased by inhibition of Wnt/β-catenin signaling (102).
3.15. Sonic Hedgehog
Hedgehog (Hh) signaling pathway that activates by sonic hedgehog (Shh) has the potential to push cells toward tumorigenesis (103). The Hh pathway promotes mesenchymal markers and restrains epithelial markers based on the up-regulation of snail (104) and twist (105). In pancreatic cancer, the Hh pathway abolishes E-cadherin expression and increases N-cadherin and snail expression (106). Shh receptor is composed of tumor suppressor protein patched (Ptc) and smoothened (Smo). In the off Hh signaling pathway, Ptc binds to Smo and inhibits its activation. Therefore, GSK3βs phosphorylate glioma-associated oncogene homologs (Glis) to repress its protein level. When Shh binds to its receptor, Ptc became internalized for degradation. Therefore, Smo is released and activated by adding phosphate groups. Accumulation of Smo activates Gli that translocate to the nucleus for up-regulation and down-regulation expression of targeted genes (107).
4. Conclusions
Besides therapeutic progression, metastasis is still the main problem of cancer. Different parts of the metastasis mechanism have been known, but there are valuable parts that are unmapped during studies. One of the main problems is that different cells and factors have effects on metastasis. TME cells induce metastasis through secretion factors to activate signaling pathways that activate TFs for increasing expression levels of mesenchymal markers and diminishing expression levels of epithelial markers. Different cytokines trigger multiple signaling pathways in cancer types to induce EMT and push tumor cells toward metastasis. Discovering and the manifestation of the cells and their effects are a challenge. In addition, tumor heterogeneity and diversity in TME cells are a blocker for known better metastasis and gain to cancer treatments. Co-culturing different TME cells with tumor cells to identify their effect on metastasis has been elucidated in different aspects of the mechanisms. Design a model with multiple TME cells that are connected to primary tumor mass is an option to make progress for manifestation the metastasis. However, the combination of different TME cells to evoke metastasis has a small part in the literature. In addition, the therapeutic potential of TME cells in metastasis needs more attention to find a solution for inhibition of metastasis.
![Secreted factors and signaling pathways are involved EMT [modified (<a href="#A113121REF61">61</a>)] Secreted factors and signaling pathways are involved EMT [modified (<a href="#A113121REF61">61</a>)]](https://services.brieflands.com/cdn/serve/3170b/52813bdd3492f74d95033d3ad7b234456631623c/ijcm-113121-i001-F1-preview.webp)