1. Background
Head and neck malignancy has been ranked as the sixth most frequently diagnosed cancer and laryngeal cancer (LC) is the most frequently observed member of this family and represents the 11th most prevalent and deadliest cancer in the world. The incidence of this disease in respiratory neoplasms is in the second place (95%) (1). LC occurs at any mucosal surface of the larynx and is the deadliest type of cancer. (2). According to the evidence, the 5-year survival rate of LC is 60%. More than half of the patients (54%) are diagnosed and cured before metastasis of the tumor and expansion of malignant cells outside the larynx, which increases the 5-year survival rate to 77%. In contrast, the 5-year survival rate will be less than 45% if the tumor has extended to nearby tissues or lymph nodes. The 5-year survival rate in patients with metastasis is 33%. However, the site of the tumor (subglottis, supraglottis, glottis) is also another factor that influences the 5-year survival (3). The most prevalent pathological type of laryngeal malignancy is squamous cell carcinoma (LSCC). According to the results of recent studies, the rate of LC is increasing. Even though considerable advances have occurred in surgery and radiation therapy during the past decades, the 5-year survival rate of LSCC has not improved significantly to the high local recurrence rate. The main treatments for LC are surgery, radiotherapy, chemotherapy, and combination therapy (4). Therefore, for early diagnosis of LC at all stages, even at primary stages whose clinical symptoms are concealed, recognition of greatly sensitive biomarkers seems to be essential for the amelioration of LC patients' outcomes.
MicroRNAs are a group of molecules that has recently become an interesting field of research that may be followed by the recognition of new biomarkers and new therapeutic targets. Consequently, highly specific and sensitive biomarkers are required for the prognosis prediction and early diagnosis of LC even in the early stages without clinical symptoms, and new therapeutic agents are considerably needed to be more effective for controlling and targeting LC cells. This gene family transcribes as a small non-encoding RNA of 22 nucleotides and, then, binds to the complementary mRNA leading to translational suppression or transcriptional degradation. miRNAs play a clear role in different kinds of human malignancies (5). In many diseases, the miRNA expression profile shows marked changes, which are particularly evident in tumors (6). miRNA expression analysis is helpful in the early detection and evaluation of tumor prognosis, metastasis, recurrence, and diagnosis in different malignancies (7). Early and accurate diagnosis in the different stages of the malignancy is important to reach a high survival rate in LSCC; however, early diagnosis is often inefficient due to the lack of specific symptoms (8). For this reason, we reviewed the recent literature and selected these microRNAs, which are studied as prognostic and diagnostic markers in LSCC and other cancers.
2. Objectives
In this research, we aimed at investigating the expression profile of 4 human miRNAs in 30 LC tissues in comparison to adjacent healthy tissues, hoping to detect appropriate and potent markers.
3. Methods
3.1. Study Population
SCC tissues and marginal normal tissues were collected through the laryngectomy from 30 patients between February 2018 and 2019 at Tabriz University of Medical Sciences (Imam Reza Hospital). None of the patients had any experience with radiotherapy or chemotherapy. All samples were transferred directly to the ribonuclease inhibitor solution (QIA Gene Cat NO: 76104) and stored at -80°C until the next step. Current research has been permitted by the Ethics Committee of Tabriz University of Medical Sciences and all participants read and signed written consent (Ethical code: IR.TBZMED.REC.1398.999). The general characteristics of the participants are shown in Table 1.
| Variables | Number of Cases |
|---|---|
| Age (y) | |
| < 55 | 16 |
| ≥55 | 14 |
| Gender | |
| Male | 19 |
| Female | 11 |
| Tobaccoexposure | |
| Smoker | 18 |
| Nonsmoker | 12 |
| Differentiation | |
| Well | 19 |
| Moderately/poorly | 11 |
| Clinicalstage | |
| I/II | 17 |
| III/IV | 13 |
| Lymph node metastasis | |
| Negative | 17 |
| Positive | 13 |
| Distant metastasis | |
| Negative | 20 |
| Positive | 10 |
3.2. Total RNA Isolation, Reverse Transcription, and Quantitative PCR
Total RNA extraction from tissues was done by the manufacturer's protocol, using TRIzol reagent (Roche Cat NO: 11667165001). Afterward, the Nanodrop tool was used to check the quantity and quality of RNA (Thermo Fisher Scientific, USA). After that, the samples were stored at -80°C until the next step. For cDNA synthesis, the stem-loop method and 2x RT-PCR (Taq) pre-mixture kit (BioFACT, Seoul) were used. The gene expression level was determined by a Step-one Real-time PCR device (Applied Biosystems, USA) and SYBR Green Master mix (Takara, Korea). In addition, the housekeeping gene for the normalization of miRNA expression was U6. Formula 2-ΔCT was used to calculate the relative miRNA expression (Table 2).
| Micro-RNA | Stemloop | F Primer | R Primer |
|---|---|---|---|
| miR-126-5P | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCCTA | CGTGCTCATTATTACTTT | CCAGTGCAGGGTCCGAGGTA |
| miR-132-5P | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAAGTTTC | CGTGCTACCGTGGCTTTC | CCAGTGCAGGGTCCGAGGTA |
| miR-205-5P | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGACT | CGTGCTTCCTTCATTCC | CCAGTGCAGGGTCCGAGGTA |
| miR-302-5P | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAAGCAAG | CGTGCTACTTAACGTGG | CCAGTGCAGGGTCCGAGGTA |
| U6 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAATAT GCTTCGGCAGCACATATACTAAAAT | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
3.3. Statistical Analysis
Statistical analysis was carried out, using Graph Pad Prism v.6.00 software to examine the gene expression panel of microRNAs in cancer and marginal samples based on the unpaired t-test program. ROC curve analysis was used to assess the potentiality of each of the microRNAs as a biomarker. The value of P < 0.05 was considered significant and all values have been defined as mean ± standard deviation.
4. Results
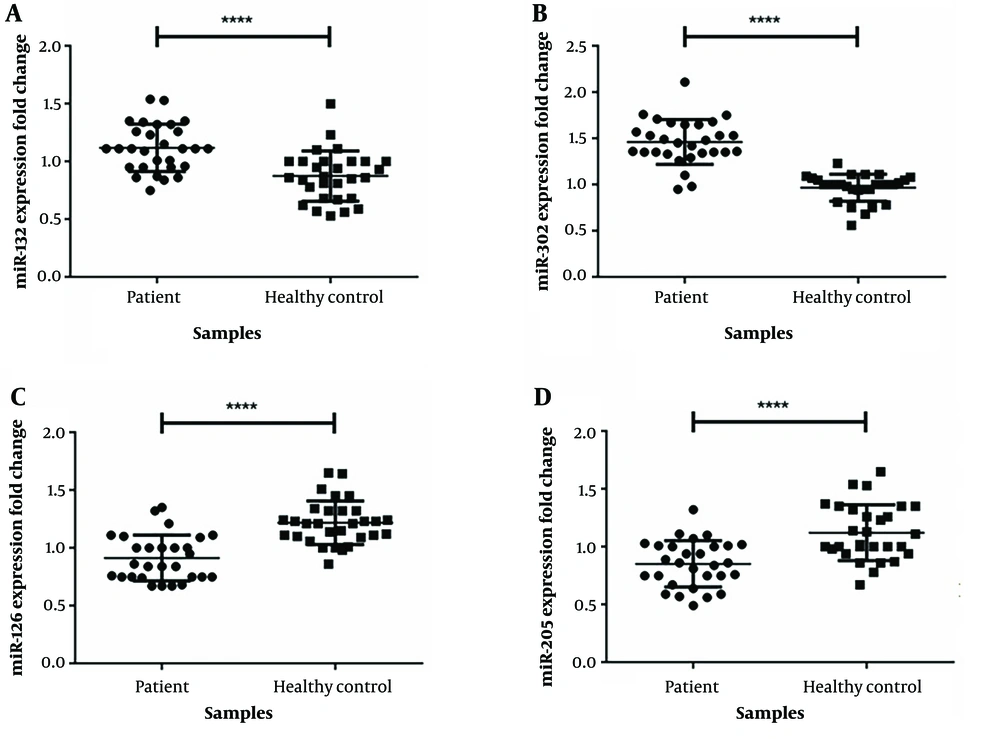
4.1. miR-302 and mirR-132 are Significantly Up-regulated in Laryngeal Squamous Cell Carcinoma Cancer
Our data show significant overexpression for miR-132 (fold change: 1.28, P < 0.0001; Figure 1) and mirR-302 (fold change: 1.05, P < 0.0001; Figure 1) in tumor cells compared to marginal normal cells. Analysis of the association between miR-302 and miR-132 expression and clinical-pathological features of patients had a significant connection among miR-302 and miR-132 expression and different tumor stages in the samples (P < 0.0001) (Table 3).
| Micro-RNAs | Age | Sex | Tobacco Exposure | Differentiation | Clinical Stage | Lymph Node Metastasis | Distant Metastases |
|---|---|---|---|---|---|---|---|
| P-Value | |||||||
| miR-126-5P | 0.27 | 0.21 | 0.41 | 0.067 | < 0.0001 | < 0.0001 | < 0.0001 |
| miR-132-5P | 0.098 | 0.15 | 0.23 | < 0.0001 | 0.071 | 0.067 | 0.057 |
| miR-205-5P | 0.086 | 0.45 | 0.121 | 0.079 | 0.073 | 0.053 | 0.063 |
| miR-302-5P | miR-302-5P | 0.31 | 0.097 | 0.053 | < 0.0001 | < 0.0001 | < 0.0001 |
4.2. miR-126 and miR-205 are Significantly Low Expressed in Laryngeal Tumor
Based on the result of this study, miR-205 (fold change = 0.241, P < 0.0001, Figure 1) and miR-126 (fold change = 0.251, P < 0.0001, Figure 1) are low expressed in malignancy cells compared to marginal normal samples. Also, there was a significant correlation between miR-126 expression level and tumor stages of the patients (Table 3). However, we did not find any interaction between the expression of miR-205 level and clinicopathological features of the patients.
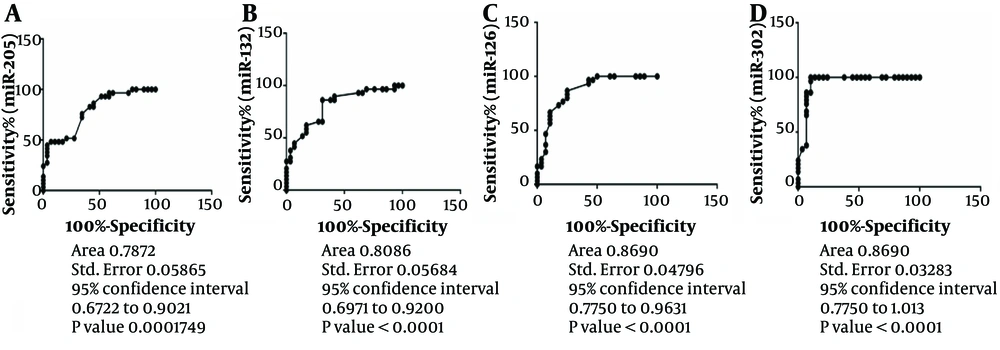
4.3. miR-302 as a Diagnosis Marker in LSCC
We used the ROC curve to evaluate miR-205, mirR-132, MiR-126, and MiR-302 specificity and sensitivity as new biomarkers in LSCC. The results determined a ROC region biomarker index of 0.7872, 0.8086, 0.8690, and 0.9489 in LSCC patients, respectively (Figure 2). Among these miRNAs, miR-302 has more specificity and sensitivity to discriminate cancer tissues from healthy marginal (Area: 0.9486) (Figure 2).
5. Discussion
LC is the most frequently diagnosed type of head and neck tumor and LSCC is the most prevalent type of LC (85%). In early-stage diagnosis, the rate of survival is about 90%, but in late-stage diagnosis, the survival rate is about 50%. So, it is critical to find biomarkers for early detection in LC (3, 9). miRNAs could be a precise and helpful biomarker for early diagnosis in tumors including LC (10). The miRNAs, selected for our study, were dysregulated in different types of malignancy and are involved in the pathogenesis of different malignancies including LSCC. The expression level was investigated hoping to identify appropriate biomarkers for LSCC. In this study, miR-302 and mirR-132 were considerably overexpressed, and miR-205 and miR-126 were considerably low expressed in LSCC tissues. As far as we know, there is no such research that has been specifically performed to evaluate the expression level of miR-302a in LSCC tumors, but some previous research reported the aberrant expression of miR-302a in a variety of malignancy types. For instance, a reduction in miR-302a expression level in breast malignancy affects metastasis and invasion controlling of breast tumor cells (11). Previous research has shown that the miR302/367 cluster participates in almost all the stages of the tumor development process in several malignancies (12). Based on previous studies, the miR-302a expression correlates with necrosis and a high level of WRAP53 (13). Guo et al. revealed that miR-302a was considerably low expressed in human ovarian tumor cells, compared with the normal cells. The results suggested that changes in miR-302a expression could be involved in ovarian tumor progression. The cell growth and progression in the miR-302a transfected cells (ovarian cancer) was considerably reduced compared to the control group (14). Zhao et al. have demonstrated that the miR-302a, miR-302d, miR-302c, and miR-302b were considerably low expressed in P-glycoprotein (P-gp)-overexpressing MCF-7/ADR cells. They also revealed that miR-302a/d/b/c ectopic combination expression through targeting the P-gp gene intensifies the sensitivity of breast tumor cells to the anticancer drug PAC, VP-16 (15). Yang CM et al. have shown that miR-302/367 cluster expression in glioblastoma cells prevents the expression of the carcinogenic gene (16). Lower expression of miR-302a has a relation with the VEGF-A expression restrains cell growth and invasiveness and promotes apoptosis in HCC. On the other hand, interruption of cell growth, cell cycle suspension (in vitro), and tumor development (in vivo) in prostate cancer may be the result of miR-302 up-regulation (17). Up-regulation of the miR-302a in the colorectal tumor may prevent cancer cell progression and invasion by preventing the expression of associated proteins via controlling the MAPK and PI3K / AKT signaling pathways (18). Our findings confirm that miR-302 is more expressed in LSCC patient tissue than in normal tissue. To the best of our knowledge, this is the first time that miR-302 expression level has been studied in LC.
Currently, there is little data on the expression of miR-132 in LSCC. Lian et al. revealed that miR-132 is highly expressed in LC cells directly targets FOXO1 (tumor suppressor), and acts as an important inhibitor of PI3K/Akt signaling. This research confirmed the miR-132 tumorigenic role in LC by controlling the PI3K/AKT/FOXO1 pathway (19).
On the other hand, an in vitro study by Chen et al. on LSCC revealed that miR-132 functions as a tumor suppressor and performs a considerable role in preventing growth, and migration, enhances chemo-sensitivity, and invasion via controlling TGF-β1/Smad2/3 signals (20). In this research, we detected that miR-132 was highly expressed in LSCC cells and this overexpression was related to the differentiation in LSCC tissues. Most previous studies in various tumors have shown that miR-132 was down-regulated, but in our research, miR-132 was overexpressed (21).
Concerning miR-126, we found that the miR-126 plasma level was reduced in patients with LSCC. Also, we found that miR-126 low expression in patients with LSCC was involved in lymph node metastasis and differentiation. Xin Sun et al. confirmed that miR-126 plasma levels were decreased and could be used as a prognostic marker in patients with LSCC. In addition, they showed that miR-126 plasma levels were negatively correlated with Camsap1 expression. According to these findings, they hypothesize that Camsap1 may be a new target gene for miR-126 (22).
In another study, Sassahira et al. revealed that low expression of miR-126 is involved in tumor development through induction of angiogenesis and lymphatic angiogenesis via VEGF-A activation in the oral LSCC cell line (23). Also, Yu et al. reported that low expression of miR-126 promotes oral tumors in animal models (24). Liu et al. confirmed that miR-126 is low expressed in lung tumor tissues compared to normal lung tissue and low expression of mir-126 leads to VEGF-A up-regulation in lung malignancy cells (25). They also indicated that the miR-126 expression level was low expressed in esophageal cancer (26). Guo et al. confirmed that miR-126 has a tumor suppressor role, and there is a relation between the miR-126 down-regulation and disrupted signaling via PI3K due to the low expression of its controlling subunit p85 in colon malignancy (27). The expression of mir-205 has shown a bilateral effect in various tumor types, which depends on the stage of malignancy and cell of origin. In certain cell types, miR-205 promotes the generation and progression of tumors as an oncogenic factor. In other types, it prevents cell invasion, growth, and EMT and acts as a tumor inhibitor (28).
In the current research, the expression level of the miR-205 was lower in LSCC tissues in comparison with marginal normal tissues. Tian et al. indicated the tumor suppressor outcome of the miR-205, which may inhibit cell proliferation by regulating Bcl-2 and induce LSCC apoptosis. They also proposed miR-205 as a high-value and potential novel target for therapeutic methods in LSCC (29). Boll et al. found a lower expression level of miR-205 in prostate malignancy. They also indicated that miR-205 up-regulation suppresses major carcinogenic pathways in prostate cancer (30). Lee et al. confirmed that miR-205 low expression causes to elevation of the expression levels of ZEB1 and ZEB2 and a reduction in the expression level of the E-cadherin transcriptional, thereby inducing growth and invasion in breast tumor cells (MCF-7) (31). Also, in 2011, Matsushima found that the CDK2AP1 (tumor suppressor) gene was down-regulated by miR-205, demonstrating that miR-205 controls CDK2AP1 in esophageal squamous cell carcinoma (ESCC). Thus, miR-205 affects the malignant cells as an oncogene via suppressing CDK2AP1 and promotes cell growth and motility by boosting the expression levels of Cyclin D1, C-Myc, MMP-9, and MMP-2 in ESCC cells (32). In this study, we found a positive relationship among miR-302, miR132, and miR-126 expression levels, metastasis, and lymph node involvement. In summary, our research showed that these 4 miRNAs have the potential as a diagnostic or prognostic biomarker in case of future studies confirmation.
5.1. Conclusions
In the present research, we found that miR-302 and miR-132 overexpressed and miR-126 and 205 low expressed in LSCC tissues compared with normal marginal tissues and may be used as a prognosis or diagnosis factor in LSCC.