1. Background
The human lung adenocarcinoma cell line A549 was inaugurated by D.J. Giard in 1972 from grade IV lung cancer tissue removed from a 58-year-old Caucasian man and subsequently donated to the ATCC cell line bank by M. Lieber under accession number CCL-185TM (1). The A549 cell line is known as a hypotriploid alveolar basal epithelial cell. It exhibits a polarized monolayer adherent morphology and positive tannic acid staining for lamellar bodies. The production of lamellar bodies, the expression of IA1 and IIB6 P450 isoenzymes, and the endocytic abilities of these cells have made them a suitable model for use as a type II lung epithelial cell and for in vitro studies of the oxidative metabolism of drugs in the lung (2). Consistent with the pattern of phospholipid synthesis expected for cells responsible for lung surfactant synthesis, A549 cells synthesize lecithin with a high content of unsaturated fatty acids using the cytidine diphosphocholine pathway at both early and late culture passages. Considering that A549 cells are lung adenocarcinoma-derived cells that lack tissue architecture, tumor microenvironment, and in-situ tumor cell communication, and retain gene expression patterns similar to tumors, it is a proper model for human lung adenocarcinoma studies (3-8).
It should be noted that Lung Adenocarcinoma, which is related to non-small cell lung carcinoma (NSCLC), is the most common type of lung cancer, accounting for about 40% of all lung cancer cases (7, 9).
Previous studies on A549 cells have shown that 24% of all A549 cells have 66 chromosomes. Most of these cells have two X and two Y chromosomes. However, in 40% of the cells examined, one or both Y chromosomes were missed (1). This cell line carries a homozygous mutation at position 12p12.1 (c.34G>A/p.Gly12Ser) of the Kirsten rat sarcoma virus (K-RAS) pro-oncogene protein (3, 10-12). Due to the homozygous deletion of the p.Gln37Ter (c.109C>T) and cyclin-dependent kinase inhibitor 2 A (CDKN2A) homozygous, c.1_471del471/p.M1_*157del, A549 cell line does not express the serine/threonine kinase tumor suppressor gene (STK11/LKB1) and CDKN2A locus (8, 10, 13-15). In addition, tumor suppressor in lung cancer 1 (TSLC1) and tumor suppressor NORE1A (RASSF5) are not expressed in A549 cells (16-18). However, cyclin D, cyclin-dependent kinases 4/6(CDK4/6), retinoblastoma susceptibility gene (RB1), transcription factor E2F, mouse double minute 2 homolog (MDM2), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), epidermal growth factor receptor (EGFR), tumor protein p53 (TP53), anaplastic lymphoma kinase (ALK), MYC proto-oncogene, and phosphatase and tensin homolog (PTEN) genes are expressed in the wild type as in normal cells (7, 8, 19-27).
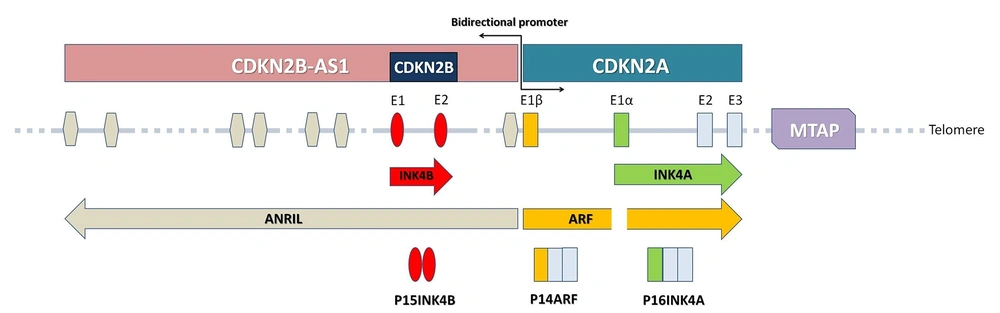
The 9p21.3 region, spanning approximately 350 kb of the genomic region, harbors 3 protein-coding genes and a long non-coding RNA in the antisense direction. The protein-coding genes consisting of S-methyl-5′-thioadenosine phosphorylase (MTAP), CDKN2A, which encodes the p16INK4A and P14ARF splice variants and cyclin-dependent kinase inhibitor 2 B (CDKN2B) (also known as P15INK4B). The long non-coding RNA in the antisense direction of CDKN2B is called CDKN2B-AS1/ANRIL (antisense non-coding RNA in the INK4 locus). The INK4-ARF locus, mapped to the 9p21.3 region and its gene cluster consisting of P14ARF, P16INK4A, P15INK4B, and long non-coding RNA (LncRNA) ANRIL. Structurally ANRIL shares a bidirectional promoter with the P14ARF gene and transcribes ANRIL in antisense orientation to the INK4-ARF gene cluster. P16INK4A is located between MTAP and ANRIL in the vicinity of the first exon of ANRIL. The first exons of P14ARF (exon1β) and P16INK4A (exon1α) are different, but the second and third exons are identical. P15INK4B is mapped to the inside of the first intron of ANRIL in antisense orientation (28) (Figure 1).
Schematic view of the INK4-ARF locus in the 9p21.3 chromosomal region. The INK4-ARF locus, mapped to the 9p21.3 region and consisting of the CDKN2A, CDKN2B and lncRNA ANRIL genes. ANRIL shares a bidirectional promoter with theP14ARF and transcribes ANRIL in antisense orientation to the CDKN2A and CDKN2B genes. P16INK4A is located between MTAP and the first exon of ANRIL. The first exons of P14ARF (E1β) and P16INK4A (E1α) are different, but the second and third exons are identical. CDKN2B is mapped to the inside of the first intron of ANRIL in antisense orientation. The A549 cell line is characterized by homozygous deletion of the CDKN2A, CDKN2B and MTAP genes.
When the cell is exposed to various stress signals such as DNA damage, oxidative stress, or enhancing oncogenes, the expression of INK4-ARF genes is activated. This results in a cascade of signaling events that adequately initiate the cell cycle arrest. P14ARF, a member of this gene cluster, interacts with the acidic domain of MDM2 to block its interaction with P53. Nuclear segregation of MDM2 intercepts MDM2-mediated delivery of P53 to the cytoplasm, thereby preventing P53 degradation. As a result, P53-dependent cell cycle arrest occurs in both G1 and G2 phases. In response to stress signals, the P16INK4A and P15INK4B are activated and attached to CDK4/CDK6 kinases. Preventing the formation of active complexes by cyclin D leads to hypo-phosphorylation of RB. Hypo-phosphorylated RB connects to the transactivation domain of E2. This complex further engages histone deacetylase 1 (HDAC1) and SUV39H1 histone lysine methyltransferase to E2F target genes, thereby hampering them and obstructing the G1-to-S phase transition (15, 29).
For the first time, the ANRIL gene was discovered through a 403 231 bp germline deletion in a French family with a history of melanoma and nervous system tumors syndrome (30). In response to genomic stress induced by DNA damage, E2F1 activates the transcription of ANRIL in an ataxia telangiectasia mutated (ATM)-dependent manner. ANRIL is transcribed by RNA polymerase II and spliced into several linear and circular isoforms in a tissue-specific manner. The complete gene has 21 exons and it has been only found in simians. Under normal circumstances, ANRIL binds to the SUZ12 subunit of Polycomb repressive complex 2 (PRC2) when DNA repair is complete. This binding helps to suppress the inhibitory effects of the INK4-ARF locus by inducing the methylation of histone 3 at lysine 27 (H3K27), allowing re-entry into the cell cycle; this has been suggested as a potential mechanism. In addition, ANRIL binds to CBX7 of Polycomb repressive complex 1 (PRC1), enabling recognition of H3K27 for monoubiquitination of histone 2A at lysine 119 (H2AK119) to maintain silencing of the INK4-ARF locus. ANRIL may regulate gene transcription through chromatin modulation (Nuclear localization hypothesis). Circular ANRIL may also be involved in post-transcriptional regulation (cytoplasmic localization hypothesis) (28, 31). Recently, genome-wide association studies (GWAS) identified the ANRIL gene as a genetic locus commonly associated with a variety of health conditions, including intracranial aneurysm, type 2 diabetes, coronary artery disease (CAD), periodontitis (PD), Alzheimer's disease, aging, frailty, glaucoma, endometriosis, multiple sclerosis, hypertension, as well as cancer (28, 32). In cancer, ANRIL-microRNA (miRNA) interactions can influence their target genes and initiate a cascade of events that leads to the intensification of the oncogenic aspect of ANRIL in proliferation, metastasis, invasion, resistance to radiotherapy, drug-induced cytotoxicity and apoptosis. This occurs through the involvement of various signaling pathways such as ATM-E2F1, PI3K/Akt, Wnt/β-catenin, NF-κB, TGF-β/Smad, Notch, and mTOR (28, 33).
Several studies have shown a homozygous deletion of the MTAP (34) and protein-coding genes of the INK4-ARF locus in the A549 cell line (7, 14, 35-38); so, the deletion of the LncRNA ANRIL in the antisense of this region is very likely from a structural point of view (Figure 1). However, the search in the database and literature did not reveal any report regarding the lack of expression of the ANRIL gene in the A549 cells, and several studies have reported the expression of ANRIL in this cell line (39-41).
2. Objectives
The presence of the LncRNA ANRIL in the A549 cell line was investigated in this study.
3. Methods
3.1. Cell Lines and Cell Culture
Human NSCLC cell line A549 was purchased from 2 cell bank centers in Iran, Pasteur Institute of Iran (Tehran, Iran) and Iranian Biological Resource Center (Tehran, Iran). The validity and authenticity of the A549 cell line were guaranteed by both of the cell banks. The human NSCLC cell line Calu-6, as well as the human normal lung fibroblast cell line MRC-5 and human hepatocellular carcinoma cell line HepG2, were acquired from the Pasteur Institute of Iran's cell bank. The cell bank of the Pasteur Institute of Iran verified and authenticated these cell lines. All cells were grown in Roswell Park Memorial Institute 1640 (RPMI1640) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin-streptomycin and maintained at 37°C in a humidified incubator with 5% CO2 (Figure 2).
3.2. Genome Preparation
The adherent cells were trypsinized and collected by centrifugation at 250 g for 5 minutes. The cell pellets were, then, resuspended in phosphate buffered saline (PBS).
The DNA was isolated from the harvested cells, using the FavorPrep Tissue Genomic DNA Extraction Mini Kit (Favorgen, Taiwan), according to the manufacturer's protocol. RNA was extracted from collected cells, using the FavorPrep Total RNA Purification Mini Kit (Favorgen, Taiwan), according to the manufacturer's recommendations and 60 µL of 0.25 U/µL DNase I (Yekta Tajhiz Aama, Iran) was used during the extraction process to eliminate any possible contamination of the genomic DNA. The samples were kept at -80°C until further processing. The quantity and quality of extracted DNA and RNA were checked, using a Nanodrop spectrophotometer (Thermo Scientific™ NanoDrop™ One, USA).
To evaluate the integrity and quality of the RNA and genomic DNA, we conducted an agarose gel electrophoresis, using 1% agarose and TAE running buffer.
Total RNA (1 µg) was reverse transcribed into cDNA, using a cDNA Synthesis Kit (catalog number: YT4500, Yekta Tajhiz Azma, Iran) containing random hexamer primers and M-MLV reverse transcriptase according to the manufacturer's instructions.
3.3. Polymerase Chain Reaction
The presence of the target sequences in obtained cDNA and genomic DNA from cell lines was checked by performing a polymerase chain reaction (PCR) reaction with specific primers (Table 1). Glucose 6-phosphate dehydrogenase (G6PD) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes were used as common isoenzyme markers and inteRNAl controls for DNA and cDNA, respectively, in all cell lines.
| Primer Name and Accession Number | Primer Sequences | Size of Product (Base Pair) | Template |
|---|---|---|---|
| P14ARF (NM_058195.4) | Forward: 5’- AGTGAGGGTTTTCGTGGTTC -3’ | 93 | DNA/cDNA |
| Reverse: 5’- AGTAGCATCAGCACGAGGG -3’ | |||
| P15INK4B(CDKN2B) (NM_004936.4) | Forward: 5’- TGGGAAAGAAGGGAAGAGTGTC -3’ | 173 | DNA/cDNA |
| Reverse: 5’- TCGCACCTTCTCCACTAGTC -3’ | |||
| P16INK4A(CDKN2A) (NM_058197.5) | Forward: 5’- AGCAGCATGGAGCCTTCG -3’ | 124 | DNA/cDNA |
| Reverse: 5’- GCCTCCGACCGTAACTATTC -3’ | |||
| ANRIL 1(NC_000009.12) | Forward: 5’- TCACCTGACACGGCCCTACC -3’ | 292 | DNA |
| Reverse: 5’- TCAGAGGCGTGCAGCGGTTTAG -3’ | |||
| ANRIL 2 (NC_000009.12) | Forward: 5’- ATGCTTTCTTTAGATCAACCCAG -3’ | 356 | DNA |
| Reverse: 5’- TACTCTGGCAAGACGGAGG -3’ | |||
| ANRIL 3 (NC_000009.12) | Forward: 5’- ATTGTCCATATCACTTAACCAGTTG -3’ | 348 | DNA |
| Reverse: 5’- TCATCACAGCAGTACAGAGGAAG -3’ | |||
| ANRIL 4 (NC_000009.12) | Forward: 5’- AATTGAAGGATCAGGGAGTCAG -3’ | 497 | DNA |
| Reverse: 5’-ATTCCCATGATTCACTGTAGGC -3’ | |||
| ANRIL 5 (NC_000009.12) | Forward: AAGTGGCAGGAATTTGGGAATG -3’ | 84 | DNA |
| Reverse: AGTCACTGGTCTGAGTTCTTAAA -3’ | |||
| ANRIL 6 (NC_000009.12) | Forward: 5’- TAATGCTTACCTAGTGCCAGATG -3’ | 165 | DNA |
| Reverse: 5’- AAATCCCAGCCAATTACCAGCG -3’ | |||
| ANRIL- RT1 (NR_003529) | Forward: 5’-AGAGAGGGTTCAAGCATCAC -3’ | 121 | cDNA |
| Reverse: 5’-TCTGATGGTTTCTTTGGAGTTAG -3’ | |||
| ANRIL- RT2 (NR_003529) | Forward: 5’- TTATTCCTGGCTCCCCTCGTC -3’ | 222 | cDNA |
| Reverse: 5’- TGTCCAGATGTCGCGTCAG -3’ | |||
| G6PD (NG_009015) | Forward: 5’- AGACGAGCTGATGAAGAGAGTGG -3’ | 174 | DNA |
| Reverse: 5’- AATGTGCAGCTGAGGTCAATGG -3’ | |||
| GAPDH (NM_001256799.3) | Forward: 5’- AAATCAAGTGGGGCGATGCTG -3’ | 192 | cDNA |
| Reverse: 5’- TGATGATCTTGAGGCTGTTGTCA -3’ |
The List of Primers for Amplification of the Target Sequences in the INK4-ARF Locus at the 9p21 Region
The PCR reaction was carried out according to the procedure of the 2X PCR master mix kit (Yekta Tajhiz Azma, Iran). The reaction mixture was as follows: Ten µL of master mix, 0.25 µL of forward and reverse primer (10 µM), 1 µL of DNA/cDNA template, and 8.5 µL nuclease-free water to a final reaction volume of 20 µL. A no-template control (NTC) was included for each primer pair.
The thermal cycle programs were as follows: Initial denaturation for 5 minutes at 94°C, followed by 40 cycles of 94°C for 20 seconds, Annealing 56 - 60°C (based on primer annealing tm) for 20 seconds, and extension at 72°C for 20 seconds. The final extension was 72°C for 5 minutes.
The PCR products were confirmed by 2% agarose gel electrophoresis, using Tris-acetate-EDTA (TAE) running buffer and visualized on a UV transilluminator.
3.4. Real-time PCR
Real-time PCR was performed to assess ANRIL expression in cDNA libraries from Calu-6 and A549 cell lines with the protocol of Smart Green qPCR (real-time) master mix (Yekta Tajhiz Azma, Iran) containing primers for ANRIL- RT1, ANRIL- RT2 (Table 1), using a LightCycler® 96 instrument (Roche, Germany) for 45 cycles as follows 95°C for 20 seconds, 58°C for 20 seconds, 72°C for 20 seconds, and post-denaturation for 3 minutes.
4. Results
4.1. All Protein-Coding Genes in the INK4-ARF Locus Are Homozygous Deleted in the A549 Cell Line
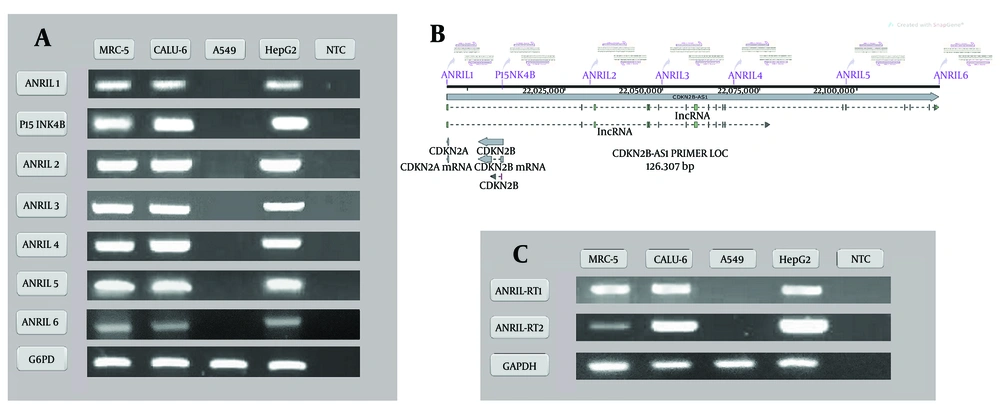
The presence of protein-coding genes located at the INK4-ARF locus at 9p21 region, including P15/CDKN2B-P16/CDKN2A-P14/ARF, was investigated at both DNA (Figure 3A and Appendices 9, 11, 13, 15 in Supplementary File contain the complete photographs of the gels) and RNA levels (Figure 3B and Appendices 10, 12, 14, 16 in Supplementary File contain the complete photographs of the gels) using specific primers. Based on our observations, the sequences of all three genes were lost in both investigated A549 cell lines. In the other examined cell lines used as control, including the normal diploid human lung cell line (MRC-5), the NSCLC cancer cell line (Calu-6), the liver carcinoma cell line (HepG2), and the sequences of all three protein-coding genes were detected at the DNA and mRNA levels.
A, the amplification bands of the protein-coding genes (P14ARF, P15INK4B and P16INK4A) located at the ink4-ARF locus at the 9p21 region in the DNA extracted from the MRC-5, Calu-6, A549 and HepG2 cell lines; B, the amplification bands of the protein-coding genes (P14ARF, P15INK4B and P16INK4A) of the INK4-ARF locus at the 9p21 region in the cDNA libraries prepared from the MRC-5, Calu-6, A549 and HepG2 cell lines. NTC, non template control. See the Supplementary Figures section for the complete photography of the gels.
4.2. ANRIL Long Non-coding RNA is Entirely Deleted in A549 Cells
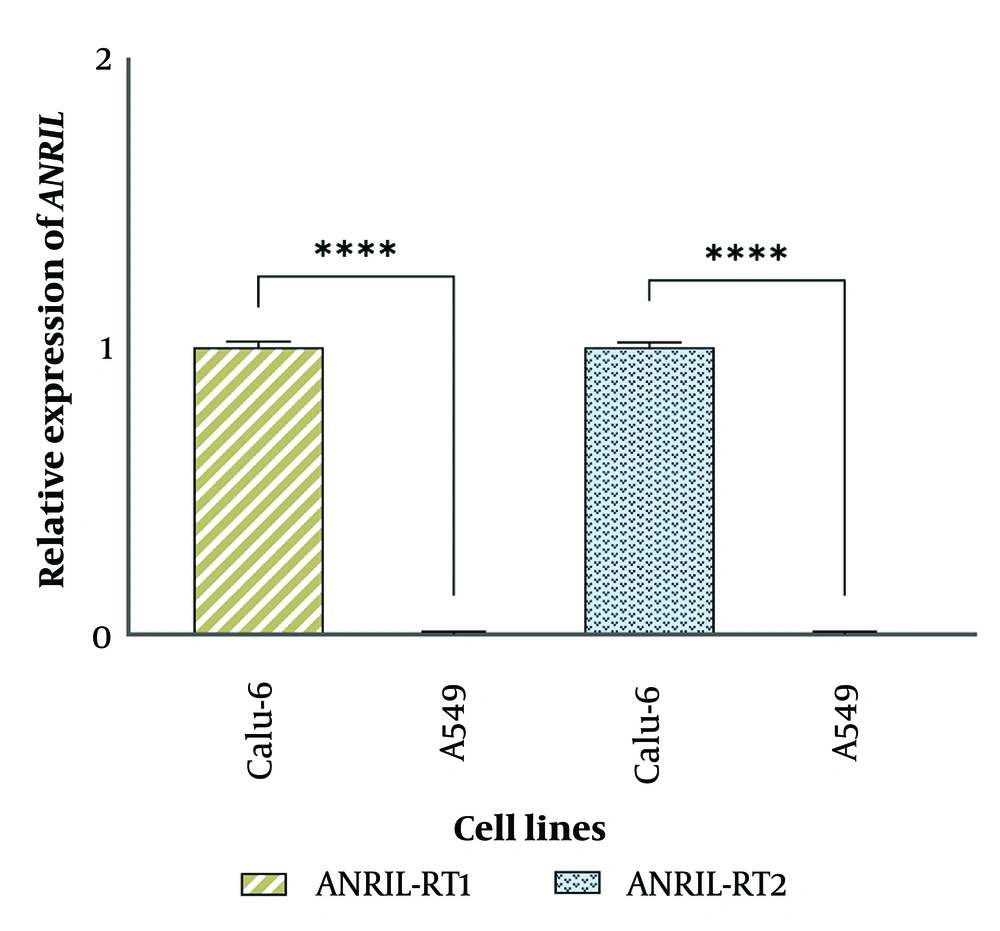
To further investigate the presence of ANRIL in the 9p21 locus, 7 regions of the entire genomic sequence of this gene (126.3 kb) were examined, using specific primers (Figure 4B). In addition, the presence of several common exons of the ANRIL gene was also evaluated at the RNA level by conventional and real-time PCR. The conventional PCR products of all the genomic regions were observed on agarose gel electrophoresis for the MRC-5, Calu-6, and HepG2 cell lines. None of these targeted regions were amplified in the A549 cell line, confirming the absence of all these genomic regions in the investigated cells (Figure 4A and Appendices 1, 11, 2 - 6, 15 in Supplementary File contain the complete photographs of the gels). In contrast to other cell lines, none of the ANRIL’s targeted exons were amplified neither targeting DNA nor cDNA library indicating that this gene has no expression in the A549 cell line (Figure 4C and Appendices 7, 8, 16 in Supplementary File contain the complete photographs of the gels). The level of ANRIL expression was also assessed by real-time PCR, where the data revealed no detectable ANRILRNA for A549 cells, while ANRIL was expressed in the Calu-6 cell line. (Figure 5 and Appendix 17 in Supplementary File). All these results suggest that ANRIL is fully deleted in A549 cells.
A, the amplification bands of 7 different regions of the entire genomic sequence of lncRNA ANRIL in the DNA extracted from the MRC-5, Calu-6, A549 and HepG2 cell lines; B, the schematic map of seven selected genomic regions of the entire genomic sequence of lncRNA ANRIL; C, the amplification bands of the lncRNA ANRIL at the INK4-ARF locus in the cDNA libraries prepared from the MRC-5, Calu-6, A549 and HepG2 cell lines. NTC, non template control. See the Supplementary Figures section for the complete photography of the gels.
5. Discussion
In this study, we examined the INK4-ARF locus in the 9p21 region by PCR at both DNA and RNA levels in the A549 cell line. We compared the results with normal human lung diploid cell lines (MRC-5), other non-small cell adenocarcinoma cell lines (Calu-6), and a cell line from a distinct cancer [human hepatocellular carcinoma (HepG2)]. Our analysis revealed that all protein-coding genes located at the INK4-ARF locus, including P15/CDKN2B, P16/CDKN2A, P14ARF, as well as the long non-coding RNAANRIL in the antisense of this locus, have fully deletion at the DNA level in the A549 cell line. Meanwhile, all of these genes are present and expressed in other cell lines that were investigated.
Since the presentation of the HeLa cell line in 1951, numerous cancer cell lines have been established and propagated, and they have been utilized in research regarding cancer biology and assessing the efficacy of anti-cancer agents, both in-vitro and as xenografts in laboratory animals (in vivo) (42).
The A549 cell line has been employed for over 5 decades as a model for type II alveolar cells, as well as an appropriate model for non-small cell lung adenocarcinoma in various studies (8).
Deletion of various encoding protein genes of the INK4-ARF locus at the 9p21 region, such as P16INK4A and P15INK4B has been reported in numerous cancers, including melanoma, glioma, lung cancer, and some leukemias, as well as cell lines linked to these cancers (43-45). In several studies, loss of heterozygosity at the 9p21 region was exhibited in 52% of NSCLC malignancies, while deletion of P16 was observed in 25% of NSCLC cases (46, 47). In a study conducted by Kraunz et al., a homozygous deletion of exon 2 of the P16 gene was reported in 34% of NSCLC samples, resulting in complete P16 protein deletion in more than half of these cases. According to their findings, this deletion is due to epigenetic silencing of 2 key genes involved in DNA double-strand break repair, the Fanconi anemia complementation group F (FancF) and Breast cancer type 1 susceptibility protein (BRCA1) genes (48). In another study, 58% of NSCLC tumors examined had an abnormality in the P16 gene, and homozygous deletion of this gene was reported in 48% of these cases (49). In the study by Panani et al., P16 gene deletion was identified in 8/11 squamous cell carcinoma, 5/6 adenocarcinoma, and 2/2 large cell lung cancer samples and this is a common finding in all subtypes of NSCLC (50). Homozygous deletion of P16INK4A is known to be one of the hallmarks of the A549 cell line (14, 35, 51, 52). P15INK4B is an important protein that acts as a backup for P16 in cells. When P16 is lost, especially under stressful conditions, cells increase P15 protein levels to compensate for the loss (36). However, the simultaneous deletion of 2 P15/Pl6 genes at the D9S126 locus (9p21) has been observed in tumors in NSCLC patients and cell lines, including A549 cells (37, 38, 53, 54). P14ARF is another tumor suppressor gene co-located with P16 and P15 at the INK4-ARF locus. Except for exon 1, the other 2 exons of this gene are similar in sequence to P16. P14ARF is deleted in 19% of NSCLC primary tumors and 25% of NSCLC cell lines, inter alia the A549 cell line (7, 55).
It has been previously reported that in more than 13% of NSCLC cell lines, including the A549 cell line, the MTAP gene is homozygously deleted. The MTAP gene is located on chromosome 9p21 (34, 56, 57) Interestingly, almost all (99%) of the tumors and various cell lines that had MTAP deletion also had CDKN2A/B loss (58, 59).
A bidirectional promoter in the 5' end of the ANRIL prime exon, flanking 300 bp upstream of the transcription initiation site of P14ARF, transcribed ANRIL in the antisense orientation of the INK4B-ARF- INK4-ARF gene cluster. Therefore, it would appear that the expression of the two genes is related to each other and is influenced by E2F1 both in physiologic and pathologic conditions. It is interesting to note that the entire P15/CDKN2B-P16/CDKN2A-P14/ARF gene cluster and its transcriptional regulator gene (ANRIL) are part of 403 kb germline deletion in the French family with melanoma, the family behind the discovery of ANRIL (30, 60, 61). Similarly, based on our observations, the entire INK4-ARF locus, including the protein-encoding genes P14, P15, P16, and the long non-coding RNAANRIL were completely deleted in the A549 cells. The structural evidence indicates that the deletion of the long non-coding RNAANRIL likely occurred concurrently with the deletion of other protein-coding genes within the INK4-ARF locus.
But how possible, that the search in databases and literature did not reveal any report regarding the lack of expression of the LncRNA ANRIL gene in the A549 cell line and there are several studies indicating the expression of ANRIL in this cell line?
Another hypothesis that could be responsible for the deletion of ANRIL in the A549 cell line is the inheritance of this deletion to a population derived from a single mutated cell.
Genomic instability is one of the hallmarks of cancers. Cancer cell lines have been propagated and immortalized from cancerous tissues for use in oncology research. Over time, during in vitro culture, cancer cell lines may undergo genetic and phenotypic changes, leading to genetic heterogeneity and instability within a cell population (62).
A549 is an NSCLC cell line that homozygously expresses the endogenous KRAS G12S mutation (11, 12). A downstream effector protein in the RAS signaling pathway is the HMG box-containing protein 1 (HBP1) transcription factor. The HBP1 enhances acetylation of the INK4A promoter by facilitating the activation of the histone acetyltransferase P300 and CREB binding protein (CBP) (29). Boosting histone acetylation or inhibition of histone deacetylase activity, in turn, has been shown to induce incorrect kinetochore localization of mitotic checkpoint proteins and extend mitotic arrest (63). In addition to incorrect kinetochore localization, the extension of the mitotic process leads to increases in the possibility of errors in its various parts, such as the cell division machinery and the gene repair system, resulting in both numerical and structural chromosome abnormalities (62). It should be noted that the proximity to the common fragile region (FRA9G), the intranuclear architecture of chromatin, and the sensitivity of genomic segments of the CDKN2A locus as hotspots for DNA double-strand breaks and subsequent microhomology-mediated repair through non-homologous end joining (NHEJ) may be a major cause of homozygous deletion of INK4-ARF region (64-66). This mechanism may be a possible explanation for the occurrence of ANRIL deletion in the A549 cells.
5.1. Conclusions
Based on our observation, in addition to the protein-coding genes of the INK4ARF locus, including P14ARF, P16INK4A, and P15INK4B genes, the long non-coding RNAANRIL is completely deleted in the A549 cell line.
Two scenarios can be considered to explain the deletion of LncRNA ANRIL in the A549 cell line. First, from a structural point of view, ANRIL shares a bidirectional promoter with the P14ARF gene and transcribes ANRIL in an antisense orientation to the INK4-ARF gene cluster. P16INK4A is located between MTAP and ANRIL in the vicinity of the first exon of ANRIL. P15INK4B is mapped to the inside of the first intron of ANRIL in antisense orientation. Given that various studies have reported that the MTAP, P14ARF, P16INK4A, and P15INK4B genes are deleted in the A549 cell line and that this deletion is one of the key characteristics of this cell line for use in related studies, it appears that ANRIL was also deleted at the time of the deletion of its antisense protein-coding genes. The second scenario that could explain the deletion of ANRIL in the A549 cell line is the recent acquisition of this deletion in a population derived from a single mutated cell through successive passages under different conditions, notably considering that cancer cell lines, like cancer itself, are genetically unstable and always susceptible to acquiring new mutations.