1. Background
Acute myeloid leukemia (AML) is a lethal malignancy with rapid and unregulated proliferation of malignant cells. This uncontrolled growth is not an isolated phenomenon but is facilitated by a complex cellular network in the bone marrow microenvironment (BMM) (1, 2). Leukemic cells engage in sophisticated communication with their surrounding cells in the BMM through various mechanisms, including interleukins, small extracellular vesicles (sEVs), and other signaling molecules, highlighting the complexity of these interactions and potential therapeutic targets (3-5). The interplay between leukemia cells and mesenchymal stem cells (MSCs) within the BMM is particularly noteworthy. Leukemic cells exploit the supportive niche provided by MSCs, which, in turn, undergo significant biological alterations influenced by the leukemic cells (6). Delving into the molecular intricacies of these interactions can yield valuable insights into the mechanisms driving leukemia progression and open new avenues for targeted therapeutic interventions aimed at disrupting the supportive niche orchestrated by MSCs in the context of leukemia.
Small extracellular vesicles released by leukemic cells play an essential role in leukemia progression by mediating intricate communication within the BMM (7). These vesicles transport diverse bioactive molecules, including proteins, nucleic acids, and signaling entities, facilitating complex interactions underpinning leukemia development (8). For instance, AML-derived sEVs can facilitate the transformation of hematopoietic stem cells into leukemic cells by downregulating genes essential for normal hematopoiesis, such as CXCR4, Scf, IGF-1, CXCL12, KIT ligand, and IL-7, while concurrently upregulating genes associated with leukemia, including DKK1, IL-6, and CCL3 (9). Furthermore, sEVs modulate processes such as angiogenesis, immune response, and drug resistance within the BMM, and can serve as diagnostic biomarkers, reflecting the disease state and providing insights into the dynamic changes occurring during leukemia progression (10-13).
Since leukemic cells alter the BMM in their favor to benefit from its comprehensive support, we treated bone marrow-derived mesenchymal stem cells (BM-MSCs) with sEVs derived from AML, which are among the key mediators of the reciprocal communication between malignant cells and the niche. Upon examining the cell cycle of MSCs, we observed an increase in the G1 phase. To further assess the G1 phase, we investigated the expression of genes such as cyclins and cyclin-dependent kinases (CDKs) that are involved in advancing the cell cycle through G1, with particular attention to CCND1, CDK4, and CDK6 due to their roles in G1 progression. On the other hand, among the various signaling pathways that lead to changes in the expression of CCND1, CDK4, and CDK6, we selected the PI3K/AKT pathway and examined the expression levels of AKT1 as the primary element of this pathway, along with CCND1, CDK4, and CDK6 as its ultimate targets.
2. Objectives
This article was conducted to investigate the proliferative role of sEVs secreted from AML cells in inducing growth and proliferation and the effect on the mesenchymal stromal mesenchymal cell cycle of Niche cells.
3. Methods
3.1. Samples
The HL60 and BM-MSC cell lines were sourced from the Stem Cell Technology Research Center (STRC) in Tehran, Iran. The HL60 cell line was characterized by the presence of CD45, CD34, CD117, CD33, and CD14 markers, whereas the BM-MSC line was characterized by CD45, CD34, CD73, and CD90 markers.
3.2. Cell Culture
The initial culture of the HL60 cell line was conducted in RPMI-1640 medium (Gibco, USA), which included 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (pen/strep; Gibco, USA) within a humidified incubator maintained at 37°C with 5% CO2, until reaching 80% to 85% confluence. Following this initial growth phase, the concentration of FBS was methodically reduced over 8 days, starting with 10% for the first 36 hours, then decreasing sequentially to 7%, 4%, and ultimately 0%, each for 36 hours. After this adaptation period, the supernatant was collected and stored at -20°C for later sEVs extraction.
The BM-MSC line was cultured in DMEM medium (Gibco, USA) with 10% FBS and 1% pen/strep in a humidified incubator (5% CO2, 37°C). The medium containing suspended apoptotic cells was replaced every 48 to 72 hours. Upon reaching 80% to 85% confluence, the cells were passaged, and BM-MSCs in passages 3 to 5 were treated with AML-derived sEVs at concentrations of 20 μg/mL, 50 μg/mL, and 80 μg/mL for 24, 48, and 72 hours, respectively, with each treatment performed in triplicate. This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.RETECH.REC.1402.014).
3.3. Extraction and Characterization of the HL60 Cell Line-Derived Small Extracellular Vesicles
3.3.1. Small Extracellular Vesicles Extraction
The isolation of sEVs from the HL60 cell line supernatant was performed, using the ExoCib kit, which employs polymer deposition for the isolation process (14). A medium devoid of serum was utilized to eliminate potential interference from sEVs contained in FBS (Gibco, USA). The HL-60 supernatant was collected frozen and thawed at 4°C for sEVs extraction. After that, it was centrifuged at 3000 RPM for 10 minutes to remove cell debris; then, reagent A was added to the samples at a ratio of sample to reagent A of 5:1, vortexed for 5 minutes, and incubated for 12 hours at 4°C. Following another minute of vortexing, the mixture was centrifuged at 3000 RPM for 40 minutes at 4°C, after which the supernatant was discarded, and 50 µL of reagent B was added to the sEVs pellet per 10 cc of the initial supernatant, with the purified sEVs stored at -20°C until further use.
3.3.2. Acute Myeloid Leukemia-Derived Small Extracellular Vesicles Surface CD Markers
To evaluate the phenotypic characteristics of the sEVs, 10 mL of monoclonal antibodies against sEVs surface CD markers were incubated with 50 mL of sEVs suspension for 30 minutes at 4°C, and after washing, reading was done, using the Attune NxT flow cytometer (Thermo Fisher Scientific, USA). Calibration was achieved, using standard microbeads (0.3 - 0.1-3 µm in diameter; BD, USA) to accurately gate particles as sEVs. The sEVs were characterized, using FITC- or PE-conjugated antibodies to detect CD9, CD63, and CD81 markers.
3.3.3. Transmission Electron Microscopy
To analyze the size, shape, and distribution of AML-derived sEVs, initially, 5 mL of sEVs suspension was fixed, using 1% glutaraldehyde. Then, a mixture drop was poured on the grid coated with carbon and placed at room temperature to dry completely. Then, the grids were washed twice for 1 minute with sterile PBS. Next, the grids were stained with 1% uranyl acetate dye (PELCO, Ted Pella) for 16 minutes. Finally, the morphology and size of sEVs were analyzed, using transmission electron microscopy (TEM) (Zeiss EMloc from Germany).
3.3.4. Dynamic Light Scattering
Dynamic light scattering (DLS) was used to measure the diameter of AML-derived sEVs. The diameter of particles in 1 mL of PBS was assessed, and their absorbance was measured at 630 nm at room temperature. The results were analyzed, using ZetaSizer (V.7.12) software.
3.3.5. Bicinchoninic Acid Protein Assay
The bicinchoninic acid (BCA) protein assay was utilized to quantify the concentration of AML-derived sEVs. Following the manufacturer's instructions for the BCA kit (DNA Biotech Co, I.R. Iran), the optical density (OD) of standard serial samples (S1-S7), positive control (FBS), negative control (PBS), and test samples was measured at 595 nm, and the protein concentration of sEVs was determined based on the standard curve.
3.4. Methylthiazole Tetrazolium Test
To determine the dose of sEVs that causes maximum proliferation in BM-MSCs, methylthiazole tetrazolium test (MTT) was performed with doses of 20, 50, and 80 during 24, 48, and 72 hours. BM-MSCs (3 × 103 cells/well) were divided into two groups: Treated (test) and untreated (control). The test group received AML-derived sEVs at concentrations of 20 µg/mL, 50 µg/mL, and 80 µg/mL for 24, 48, and 72 hours in a 96-well plate, while the control group remained untreated. After the incubation period, 10 µL of MTT solution (Acros Organics, USA) was added to each well, and the plate was incubated for an additional 4 hours at 37°C. Subsequently, 100 µL of DMSO (Sigma Aldrich, USA) was added to dissolve the formazan, and the OD was measured at 570 nm, using an ELISA reader (Biotek Elx 800, USA).
3.5. Cell Proliferation Assay by Ki-67 Staining
The impact of sEVs on BM-MSC proliferation was evaluated, using the Ki-67 antigen, a marker for cell proliferation. Test BM-MSCs were exposed to 50 µg/mL of AML-derived sEVs for 48 hours, whereas control BM-MSCs were untreated. After washing, 10 µL of Ki-67 FITC antibody was added to the cells, followed by a 30-minute incubation at room temperature. The cells were, then, analyzed, using flow cytometry, and the data were processed, using FlowJo software (v.10.10).
3.6. Cell Cycle Assay
The effect of AML-derived sEVs on the BM-MSC cell cycle was investigated, using flow cytometry. Test BM-MSCs were treated with 50 µg/mL of AML-derived sEVs for 48 hours, while the control group remained untreated. After washing with PBS and adding 50 µL of cold PBS, the cells were fixed with 1 mL of cold 70% ethanol. The cells were, then, centrifuged, the ethanol was removed, and 1 mL of MASTER MIX PI solution was added. The cells were incubated at room temperature for 30 minutes and analyzed, using flow cytometry. Results were interpreted, using FlowJo software (v.10.10).
3.7. Apoptosis Analysis by Flow Cytometry
To determine the impact of sEVs on BM-MSC apoptosis, cells in the test group were exposed to treatment with sEVs derived from AML at a concentration of 50 µg/mL for 48 hours, while BM-MSCs in the control group were untreated. The experimental treatment was executed in triplicate after a 24-hour incubation period at 37°C. To assess apoptotic cells, both the supernatant and cells were collected and subjected to centrifugation at 1500 rpm for 5 minutes. The resulting cell pellet was, then, resuspended in Annexin V-X1 buffer. To prepare flow cytometry analysis, 96 µL of the cell suspension was transferred to a flow cytometry tube, adding 1 µL of conjugated Annexin V-FITC and 12 µL of propidium iodide solution. This mixture was incubated in the dark on ice for 10 minutes and diluted to a final volume of 250 µL with Annexin V-X1 binding buffer. The samples were, then, analyzed, using a flow cytometer, and data acquisition and interpretation were performed, using FlowJo software.
3.8. RNA Extraction, cDNA Synthesis & RT q-PCR Assay
Total RNA was extracted from BM-MSCs treated with 50 µg/mL of AML-derived sEVs for 48 hours and from control BM-MSCs, using Ribo EX general (GeneAll, Republic of Korea) according to the manufacturer's instructions. RNA quality and quantity were assessed by agarose gel electrophoresis and a nanodrop instrument (Thermo Fisher, USA). cDNA synthesis was performed, using the kit protocol (AddBio, Korea) with GAPDH as the housekeeping gene.
RT q-PCR was conducted, using the ABI StepOnePlus system (Thermo Fisher Scientific, USA) and SYBER Green real-time PCR master mixes (Ampliqon, Denmark). The primers for the target genes, specifically AKT1 from the PI3K/AKT pathway, as well as Cyclin D1, CDK4, and CDK6, which are critical downstream targets of this pathway and are instrumental in the progression of the G1 phase of the cell cycle, were designed, using advanced tools such as NCBI Primer-BLAST, Oligo Analyzer, and Allele ID software. Subsequently, employing the PCR instrument (Thermo Fisher Scientific, USA) and the temperature gradient method, the optimal temperature for primer annealing was determined. The alteration in gene expression was, then, calculated as the fold change using the 2-ΔΔCt (Livak) method.
3.9. Statistical Analysis
Data analysis was executed, using GraphPad Prism software version 9.5.1 (California, USA), and results were expressed as means ± standard deviation (SD). A one-way analysis of variance (ANOVA) was employed to analyze variables across multiple conditions, followed by Tukey's test for post-hoc comparisons. Further, Tukey's test was applied for post-hoc analysis. All assays were conducted independently in triplicate to ensure the reliability and reproducibility of the findings. Statistical significance was determined at a threshold of P < 0.05.
4. Results
4.1. Characterization of Bone Marrow Mesenchymal Stromal Cells and HL60
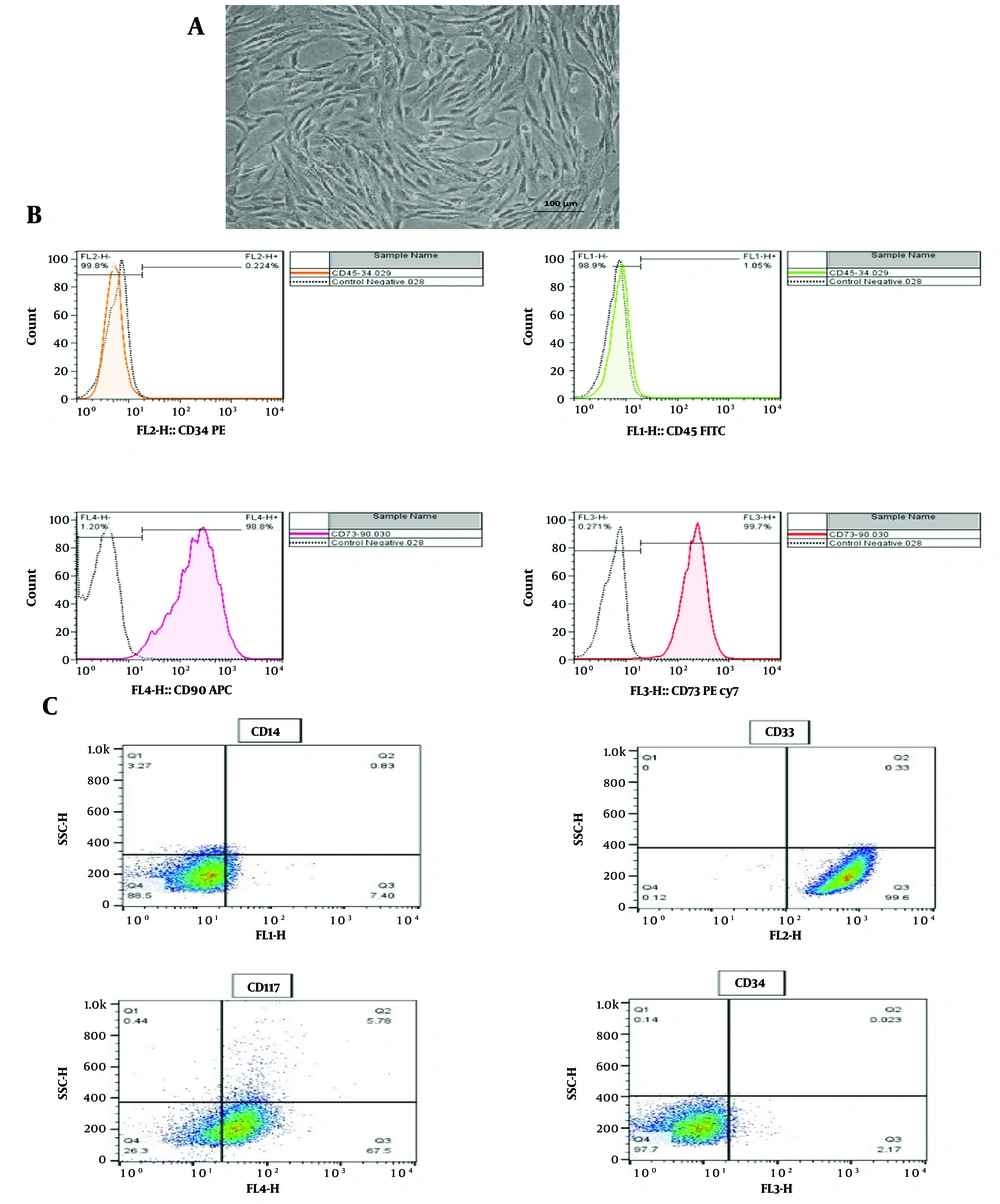
The morphological characteristics of BM-MSCs and their surface expression of CD markers were evaluated, using an inverted microscope and flow cytometry, respectively. The spindle-shaped morphology of attached BM-MSCs observed under the inverted microscope (Figure 1A), combined with the positive expression of CD73 and CD90 and the negative expression of CD45 and CD34 in flow cytometric analyses (Figure 1B), confirmed the identity of the BM-MSCs. In addition, flow cytometric analysis of the HL-60 cell line revealed the dim expression of CD45, negative expression of CD14 and CD34, and positive expression of CD33 and CD117, which conclusively indicated the myeloid origin of this cell line (Figure 1C).
Characterization of BM-MSCs and HL-60 cell line. A, spindle-shaped morphology in attached BM-MSCs observed through an inverted microscope; B, flow cytometric evaluation of BM-MSCs showing the expression of CD73 and CD90, with the absence of CD45 and CD34 expression; C, flow cytometric assessment of HL-60 cell line showing dim expression of CD45, negative expression of CD14 and CD34, and positive expression of CD33 and CD117. BM-MSCs, bone marrow mesenchymal stromal cells; CD, cluster of differentiation.
4.2. Identification of Acute Myeloid Leukemia-Derived Small Extracellular Vesicles
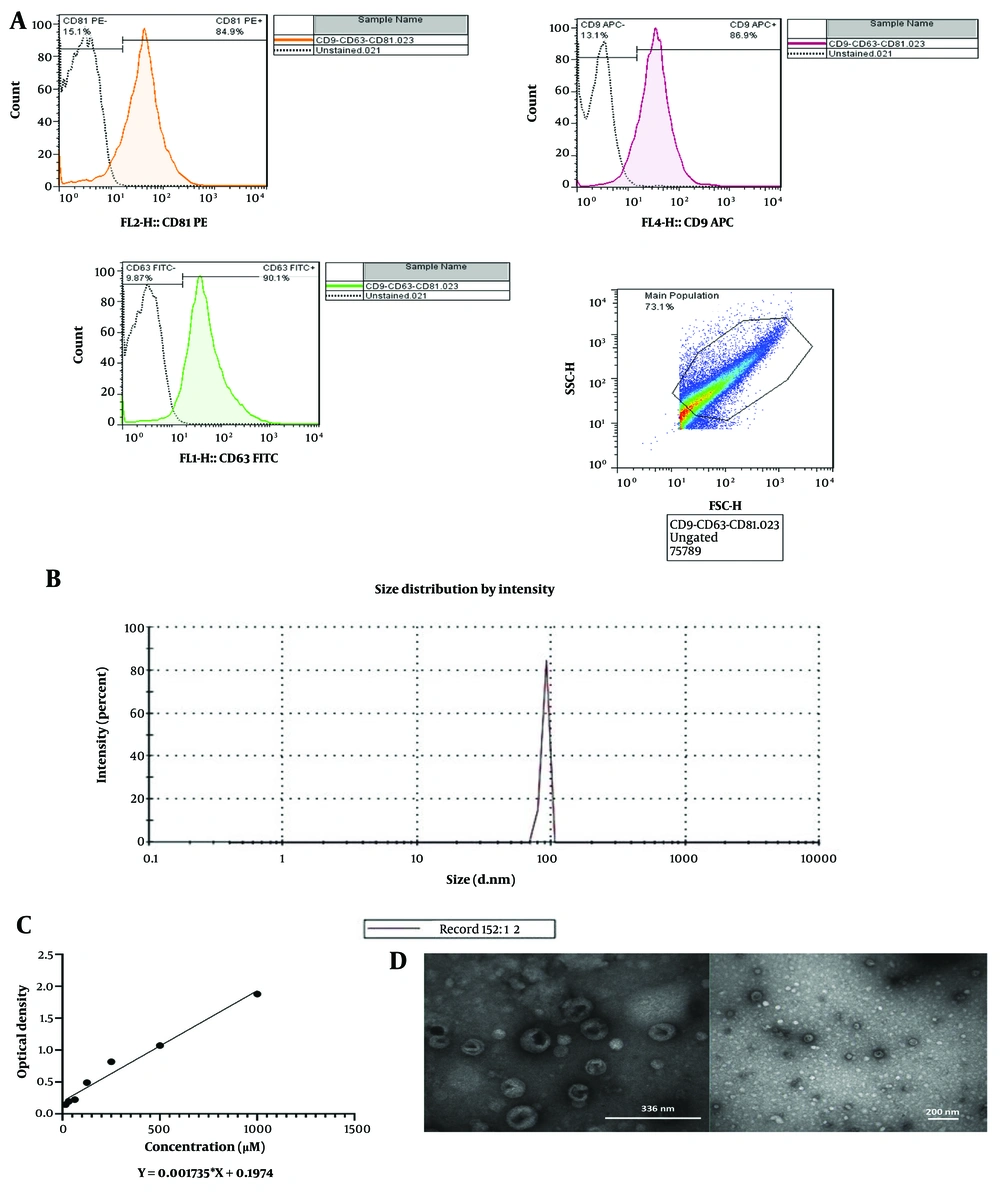
The characterization of sEVs derived from the HL60 cell line was validated, using flow cytometry, DLS, TEM, and the BCA protein assay. Flow cytometry confirmed the presence of sEVs in the harvested supernatant by detecting the surface expression of sEVs pan markers CD9, CD63, and CD81 (Figure 2A). Dynamic light scattering analysis revealed an average sEVs size of 89.36 nm in diameter (Figure 2B). The BCA protein assay quantified the concentration of AML-derived sEVs at 1499 μg per 1000 mL (Figure 2C). Furthermore, TEM analysis illustrated the morphology of AML-derived sEVs, showing them as two-layered spheres (Figure 2D).
Identification of acute myeloid leukemia (AML)-derived small extracellular vesicles (sEVs). A, surface expression of pan-sEVs markers (CD9, CD63 and CD81) by flow cytometry; B, average size of sEVs by DLS; C, quantification of sEVs by BCA; D, two-layered spheres of sEVs with TEM. DLS, dynamic light scattering; BCA, bicinchoninic acid; TEM, transmission electron microscope.
4.3. Methylthiazole Tetrazolium Test Assay
Test BM-MSCs were incubated with sEVs concentrations of 20 μg/mL, 50 μg/mL, and 80 μg/mL for 24, 48, and 72 hours, while the control group was left untreated. The results demonstrated that the effects of AML-derived sEVs were both dose- and time-dependent. Specifically, an increase in BM-MSC proliferation was observed at 20 μg/mL after 24, 48, and 72 hours and at 50 μg/mL after 24, 48, and 72 hours, with the highest proliferation rate occurring at 50 μg/mL during the 48-hour treatment period (P < 0.0001). In contrast, the 80 μg/mL concentration decreased metabolic activity at 24, 48, and 72 hours, indicating apoptosis in the test group compared to the control group (Appendix 1 in Supplementary File).
4.4. Acute Myeloid Leukemia-Derived Small Extracellular Vesicles Increased Ki-67 Levels in Treated Bone Marrow Mesenchymal Stromal Cells
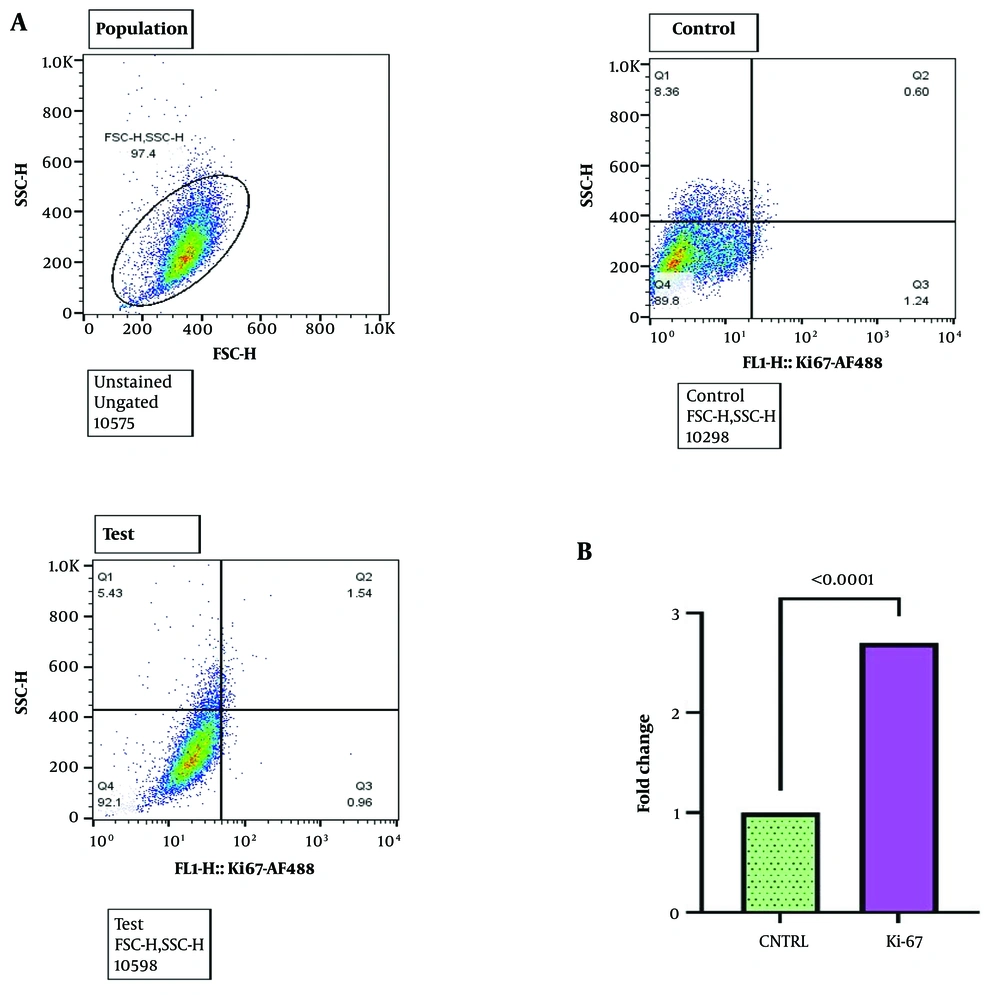
Treating BM-MSCs with AML-derived sEVs at a concentration of 50 μg/mL for 48 hours resulted in a significant elevation in Ki-67 levels compared to the control group. This was confirmed by flow cytometry analysis, which demonstrated a 2.7-fold increase (P < 0.0001) and was further supported by RT-qPCR data, showing a 2.3-fold increase in Ki-67 gene expression levels (P < 0.0001) compared to the control group (Figure 3A and B respectively).
Increased Ki-67 levels in treated BM-MSCs. A, flow cytometric analysis of BM-MSCs treated with acute myeloid leukemia (AML)-derived small extracellular vesicles (sEVs) (50 μg/mL, 48 hours) showing the population, control, and test groups. The test group exhibits a higher Ki-67 expression compared to the control; B, fold change in Ki-67 expression levels in BM-MSCs treated with AML-derived sEVs compared to the control group. The test group showed a 2.7-fold increase in Ki-67 expression levels compared to the control group. Statistical significance was assessed using paired t-test and ordinary one-way ANOVA (**** P-value < 0.0001). BM-MSCs, bone marrow mesenchymal stromal cells.
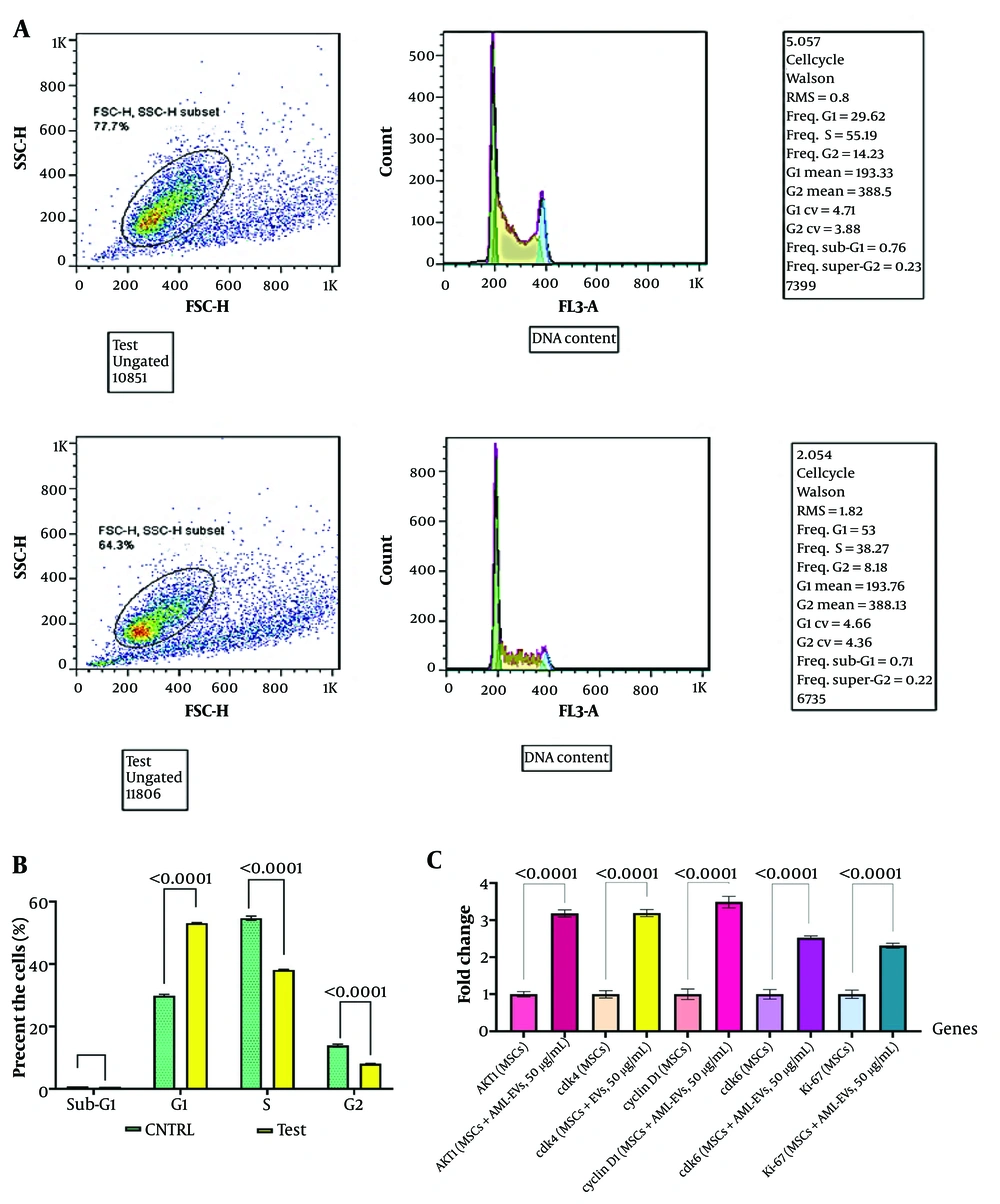
4.5. Acute Myeloid Leukemia-Derived Small Extracellular Vesicles Caused Cell Cycle Progression Through the G1 Phase
Flow cytometry analysis of BM-MSCs treated with AML-derived sEVs (50 μg/mL, 48 hours) revealed an increase in the G1 phase, showing a 1.8-fold change compared to the control group (Figure 4A and B). The RT-qPCR results for BM-MSCs treated with AML-derived sEVs (50 µg/mL, 48 hours) exhibited a significant upregulation in the expression levels of CCND1 (3.5-fold, P < 0.0001), CDK4 (3.2-fold, P < 0.0001), CDK6 (2.5-fold, P < 0.0001), and AKT1 (3.2-fold, P < 0.0001), along with Ki-67 (2.3-fold, P < 0.0001) compared to the control group (Figure 4C).
The effect of a 50 μg/mL dose of HL-60-derived sEVs on cell cycle and gene expression of BM-MSCs. A, flow cytometric analysis of BM-MSCs treated with 50 μg/mL of acute myeloid leukemia (AML)-derived sEVs for 48 hours, showing the control and test groups. The treated BM-MSCs exhibited an increase in the G1 phase compared to the control group, with a 1.8-fold change; B, quantifying cell cycle phases in BM-MSCs treated with AML-derived sEVs compared to control cells. The results show significant increases in the G1 phase and decrease in the S and G2 phases of the cell cycle (**** P-value < 0.0001); C, RT-qPCR results for BM-MSCs treated with AML-derived sEVs (50 μg/mL, 48 hours) exhibited a significant upregulation in the expression levels of AKT1, CCND1, CDK4, CDK6, and Ki-67. Statistical significance was assessed using a paired t-test and ordinary one-way ANOVA (**** P-value < 0.0001). sEVs, small extracellular vesicles; BM-MSCs, bone marrow mesenchymal stromal cells; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCND1, cyclin D1; CDK4, cyclin-dependent kinase 4; CDK6, cyclin-dependent kinase 6.
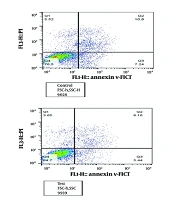
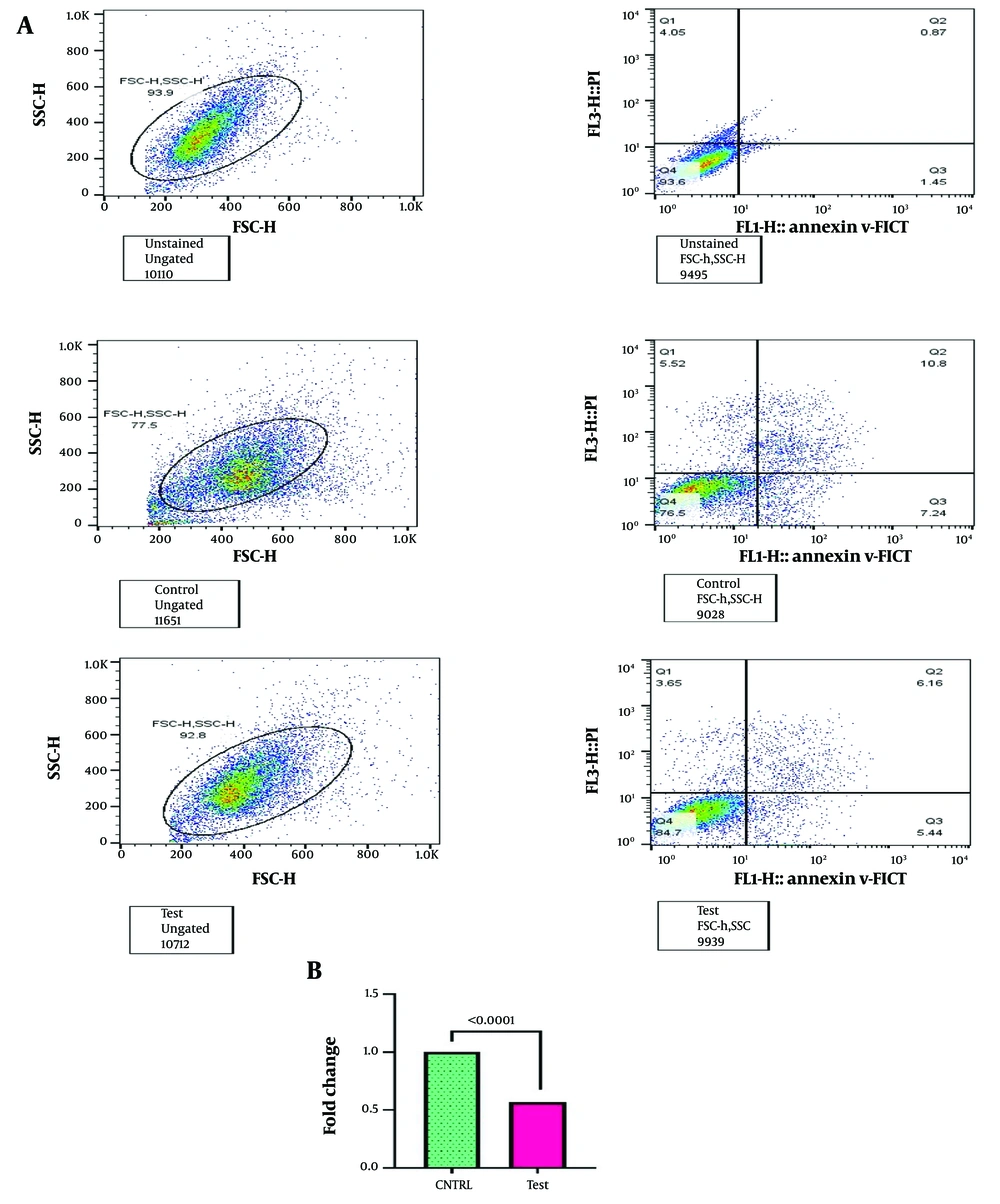
4.6. Evaluation of Bone Marrow Mesenchymal Stromal Cells Apoptosis Following Treatment with HL60-derived Small Extracellular Vesicles
The assessment indicated a significant reduction in the apoptosis rate of BM-MSCs treated with HL60 sEVs at a concentration of 50 µg/mL for 48 hours, demonstrating a 0.57-fold decrease relative to the control group (P < 0.0001), as quantified by Annexin/PI staining. This treatment resulted in a notable reduction in early and late apoptosis phases (Figure 5A and B).
5. Discussion
Acute myeloid leukemia is distinguished by its rapid onset and aggressive progression, typified by the uncontrolled proliferation of myeloid progenitor cells. Recent research has elucidated the crucial role of AML-derived sEVs in reshaping the BMM to favor leukemic cell survival and proliferation. These sEVs, laden with oncogenic proteins and RNAs, induce significant phenotypic alterations in BM-MSCs, thereby fostering AML progression through enhanced proliferation, invasion, and chemoresistance (15). The intricate cross-talk between AML cells and BM-MSCs involves sophisticated signaling pathways, notably the upregulation of S100A4 and the activation of PI3K/AKT, creating a feedback loop that sustains the malignant phenotype of AML cells (15, 16). This complex interaction promotes leukemic cell growth and impairs normal hematopoiesis, complicating therapeutic efforts. A thorough understanding of these dynamics is imperative for developing targeted therapies to disrupt these supportive interactions and improve patient outcomes. Therefore, in this study, we investigated the proliferative effects of AML-derived sEVs from the HL60 cell line on the cell cycle progression of BM-MSCs. The PI3K/AKT pathway was specifically examined due to its significant role in influencing cell proliferation through its effects on the G1 phase.
Our study demonstrated that sEVs from AML cells significantly increased BM-MSC proliferation in a dose- and time-dependent manner. This finding is consistent with several recent studies highlighting the influence of leukemia-derived sEVs on BM-MSCs. For instance, sEVs from B-cell acute lymphoblastic leukemia (B-ALL) cells have been shown to promote MSC proliferation and drug resistance (17). Similarly, sEVs from chronic myeloid leukemia (CML) cells, particularly from the LAMA84 cell line, enhance MSC proliferation and migration by promoting IL-8 secretion (18). Additionally, sEVs derived from AML cells have been reported to alter MSC behavior, enhancing their proliferation and supporting leukemia cell survival and drug resistance (6). These studies collectively underscore the critical role of leukemia-derived sEVs in modulating the BMM to favor leukemic cell survival and proliferation, providing valuable insights into potential therapeutic targets.
The increased BM-MSC proliferation in our study was evident from the MTT assay results, which showed enhanced metabolic activity of BM-MSCs treated with 50 µg/mL of sEVs for 48 hours. Further analysis revealed a 2.7-fold increase in Ki-67 levels, a cell proliferation marker, corroborated by flow cytometry and RT-qPCR data. Ki-67, renowned for its pivotal role in regulating the cell cycle and its correlation with cellular proliferation, is the most extensively employed marker for proliferation (19). Our results showed a significant increase in Ki-67 levels, measured through flow cytometry, alongside a concurrent rise in Ki-67 gene expression levels, as indicated by RT-qPCR data. Recent research has highlighted the significance of Ki-67 in understanding the biology of cancer. For example, sEVs-derived FZD10 has been identified as a crucial factor in boosting Ki-67 expression, particularly in colorectal and gastric cancer (GC) contexts, by activating phospho-ERK1/2 (20).
Similarly, Wang et al. explored the diagnostic and prognostic potential of EVs positive for the epidermal growth factor receptor (EGFR) in glioma. Their findings demonstrated the ability of EGFR in serum EVs to distinguish between high-grade and low-grade glioma patients, revealing a positive correlation with Ki-67 expression in tumor tissues (21). In a more recent study, Zadka et al. conducted a recent investigation on colorectal cancer (CRC), which found a significant positive association between Ki-67 and particular sEVs markers (CD9 and CD63) (22).
The regulation of cell cycle progression in the G1 phase is governed by intricate signaling pathways, where active biomolecules and growth agents (GA) serve as pivotal initiators. The GA-induced activation of the PI3K pathway subsequently triggers AKT1, which modulates cell cycle progression by influencing critical components, including CCND1, CDK4, and CDK6 (15). To rigorously investigate this hypothesis, we examined the ultimate targets of these signaling pathways, specifically CCND1, CDK4, and CDK6. In this study, our findings revealed a marked upregulation in gene expression associated with the G1 phase regulation, as evidenced by RT-qPCR analyses, further corroborated by flow cytometric validation. The data demonstrate that AML-derived sEVs profoundly influence the cell cycle dynamics of BM-MSCs, mainly through modulation of the G1 phase. The observed significant changes in cell cycle distribution and the upregulation of key genes within the PI3K/AKT pathway indicate that these sEVs activate pathways crucial for cell proliferation and survival. The enhanced expression of genes such as CCND1, CDK4, and CDK6 suggests that AML-derived sEVs facilitate cell cycle progression and foster a proliferative microenvironment. These findings underscore the pivotal role of sEVs in reshaping the BMM, thereby supporting leukemic cell survival and proliferation.
Moreover, Matsumoto et al. conducted a study, where they extracted sEVs from human esophageal squamous cell carcinoma (ESCC) cells, using ultracentrifugation. Employing cell proliferation assays and fluorescence imaging of the cell cycle, the investigation aimed at elucidating phenotypic changes induced by high concentrations of tumor-derived sEVs. The results, revealing a significant increase in the ratio of G1-phase cells in the sEVs exposure group, contribute valuable insights into the influence of these sEVs on cell cycle dynamics (23). This intricate interaction presents potential therapeutic targets within the sEVs-mediated signaling pathways, offering promising avenues for improving treatment outcomes in AML. The alterations in gene expression profiles strongly imply further involvement of the PI3K/AKT signaling pathways, reinforcing the critical impact of AML-derived sEVs on BM-MSC behavior and leukemia pathobiology.
Consistent with our findings, mounting evidence indicates that the sEVs-activated PI3K/AKT pathway plays a crucial role in developing and progressing various malignancies, including cervical, breast, prostate, colorectal, and lung cancers (24-30). The clinical relevance of the PI3K/AKT/mTOR pathway in cervical cancer has been highlighted by Zhang et al., who analyzed neoplastic tissues, adjacent normal tissues, and sEVs from vaginal secretions. They found no significant difference in the expression levels of PI3K/AKT/mTOR genes between cancer tissues and sEVs; however, gene expression in both was significantly elevated compared to normal marginal tissues (25). Moreover, Zheng et al. demonstrated that exosomal apolipoprotein E (ApoE) activates the PI3K/AKT pathway, facilitating the migration of GC cells (31). Another study revealed that exosomal LncRNA MALAT1 sequesters miR-26a/b, promoting the invasive behavior of CRC by activating the PI3K/AKT/mTOR signaling pathway (32).
5.1. Limitations and Suggestions
Given the involvement of sEVs and their pan markers, including CD9, CD63, and CD81, in various biological processes such as aging and fertilization, it is conceivable that suppressing these markers could give rise to significant complications. Consequently, there is a critical imperative for identifying and developing specific inhibitory tools against tumor-derived sEVs.
5.2. Conclusions
The findings of this study showed that AML-derived sEVs can further activate PI3K/AKT signaling pathways and influence the survival and proliferation of cancer cells. Our investigations suggest their specific targeting as a potential therapeutic strategy against cancer progression, invasion, and metastasis.