1. Context
Breast cancer as a complex disorder is a field of interactions between several genetic and environmental factors (1). Numerous association studies have been conducted to assess the influence of single nucleotide variants (SNPs) in human genome on susceptibility to breast cancer (2). Among possible susceptibility, loci is the gene coding for BRCA1-interacting protein 1 (BRIP1), which is a member of the DEAH helicase family (3). The essential role of BRIP1 in DNA repair and genomic stability as well as its co-localization with BRCA1 in nuclear foci (4) supports its putative role in the pathogenesis of breast cancer. Moreover, germline mutations in BRIP1 led to Fanconi anemia, a genetic disorder of both chromosome instability and cancer predisposition (5). There are a number of association studies, which have assessed the role of certain SNPs within this gene in conferring the risk of breast cancer in different populations (6-8). However, the results of these studies are inconclusive.
2. Objectives
To further examine the association between SNPs within BRIP1 and predisposition to breast cancer, we conducted a systematic search and meta-analysis of eligible association studies.
3. Data Sources
3.1. Publication Search
We searched Medline/PubMed, ISI Web of Knowledge, and Google Scholar to find eligible studies until January 2018. The search terms “BRIP1 gene”, “breast cancer”, “SNPs”, and “polymorphism” were used. In addition, we assessed the references of associated publications to get all of relevant publications. We just selected studies with full-text articles in English.
3.2. Inclusion Criteria
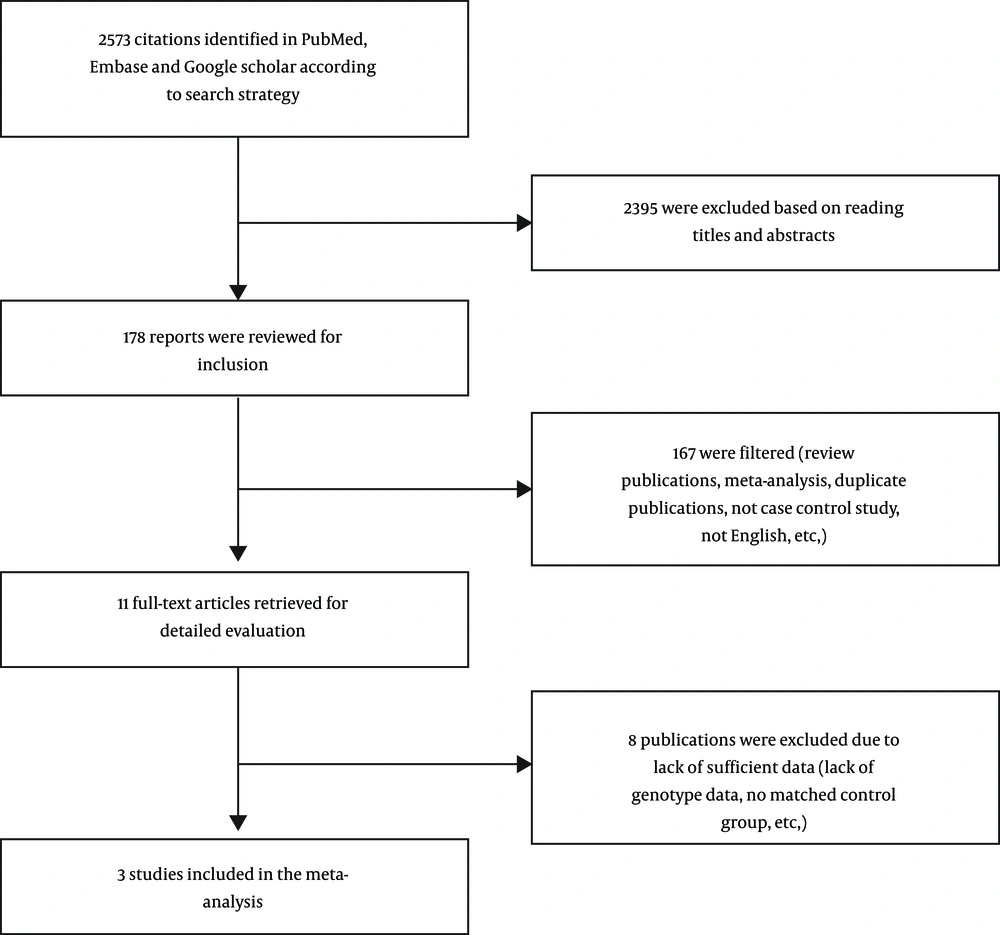
The succeeding inclusion criteria were considered: (1) Assessment of the BRIP1 variants and predisposition to breast cancer; (2) case control studies with appropriate design; (3) adequate published data to calculate odds ratios (OR) with 95% confidence interval (CI); (4) written in English; (5) reporting comprehensive genotype and alleles frequencies. Figure 1 shows the diagram of choosing studies for including in the meta-analysis.
3.3. Data Extraction
The first two authors extracted data from all eligible studies based on the mentioned inclusion criteria. First author’s family name, time of publication, origin of study, ethnicity, type of genetic material applied for analysis, total amount of cases and controls, and genotypes quantities in each study group were extracted from each selected study.
3.4. Statistical Analysis
The assessments were implemented in RevMan version 5.3 (The Cochrane Collaboration, Copenhagen, Denmark). The association between each SNP and breast cancer risk was assessed through calculation of ORs with 95% CI under 4 inheritance models, namely the allelic (wild type [W] compared with minor [M] allele), the homozygote (WW compared with MM), the dominant (WW + WM compared with MM), and the recessive (WW compared with WM + MM) models. We also assessed the compliance with Hardy-Weinberg equilibrium (HWE). The Q parameter was calculated to evaluate the amount of heterogeneity between the studies. P values greater than 0.10 were considered as indicators of homogeneity among publications. We applied the mixed-effects model and the random-effects model to merge parameters obtained from distinct studies, if they were expected to be homogenous or heterogeneous correspondingly. Funnel plots were depicted to assess publication bias.
4. Results
4.1. The Characteristics of Eligible Studies
Three publications in distinct populations including 986 cases and 1 087 controls were chosen for the meta-analysis. The rs2048718 was the only SNP that was assessed in the 3 mentioned studies. Table 1 summarizes the principle features of these publications.
4.2. Assessment of Allele Frequencies
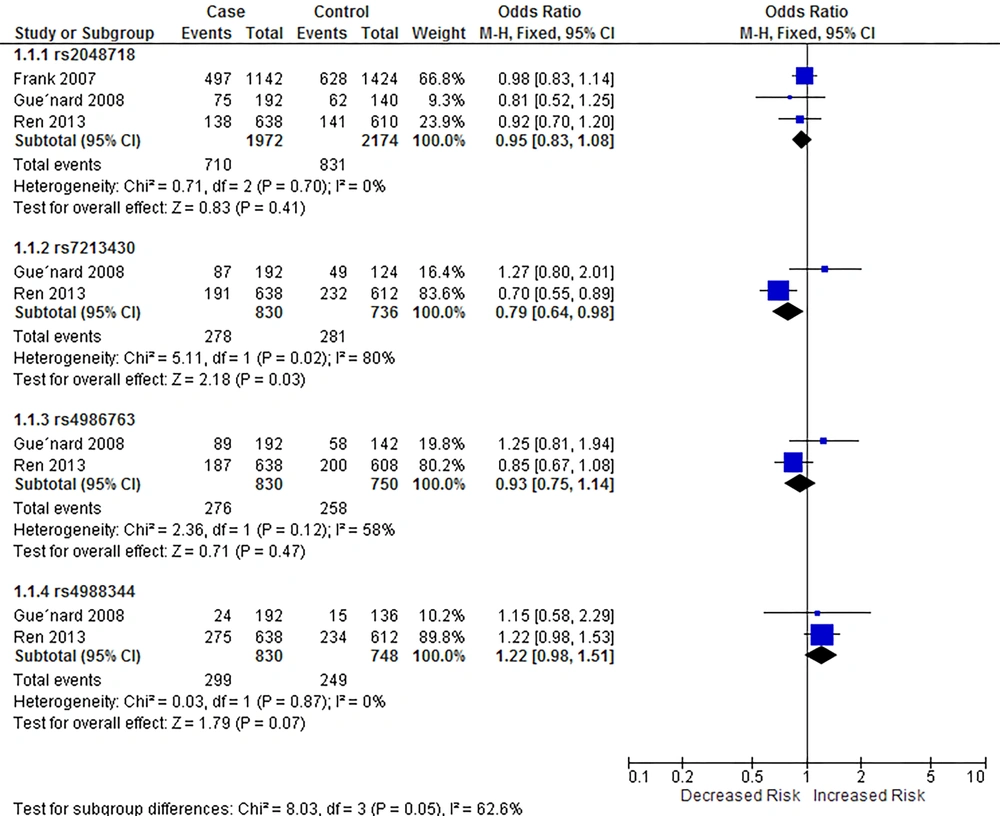
When all the studies were pooled, rs7213430 was found to be linked with breast cancer risk (OR = 0.79, 95% CI = [0.64, 0.98]). However, I2 value showed the heterogeneity between two studies included in this meta-analysis (P = 0.02). Figure 2 demonstrates the outcomes of the meta-analysis of associations between BRIP1 SNPs and breast cancer risk in allelic model.
4.3. Assessment of Genotype Frequencies
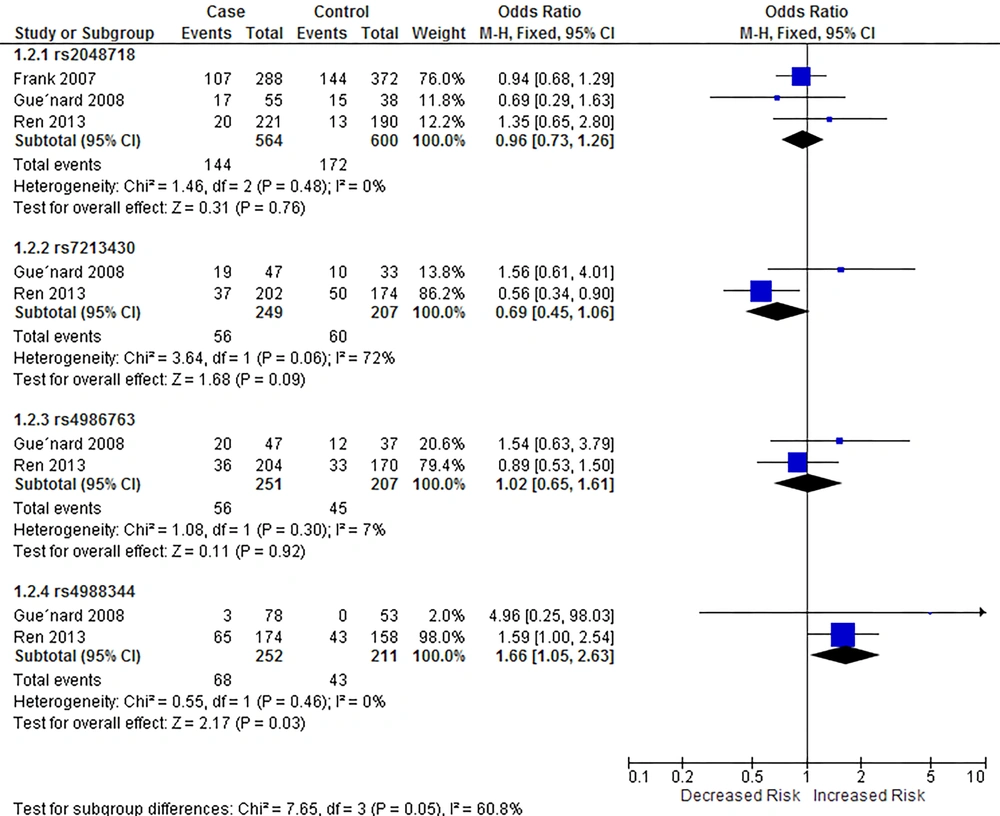
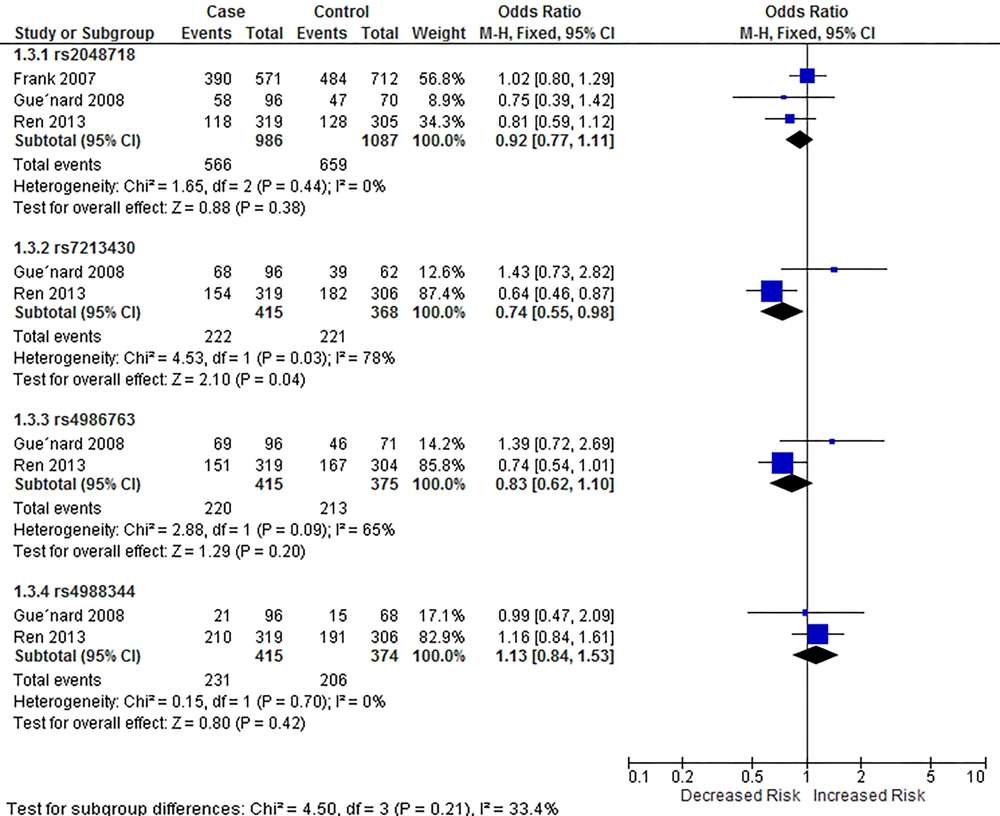
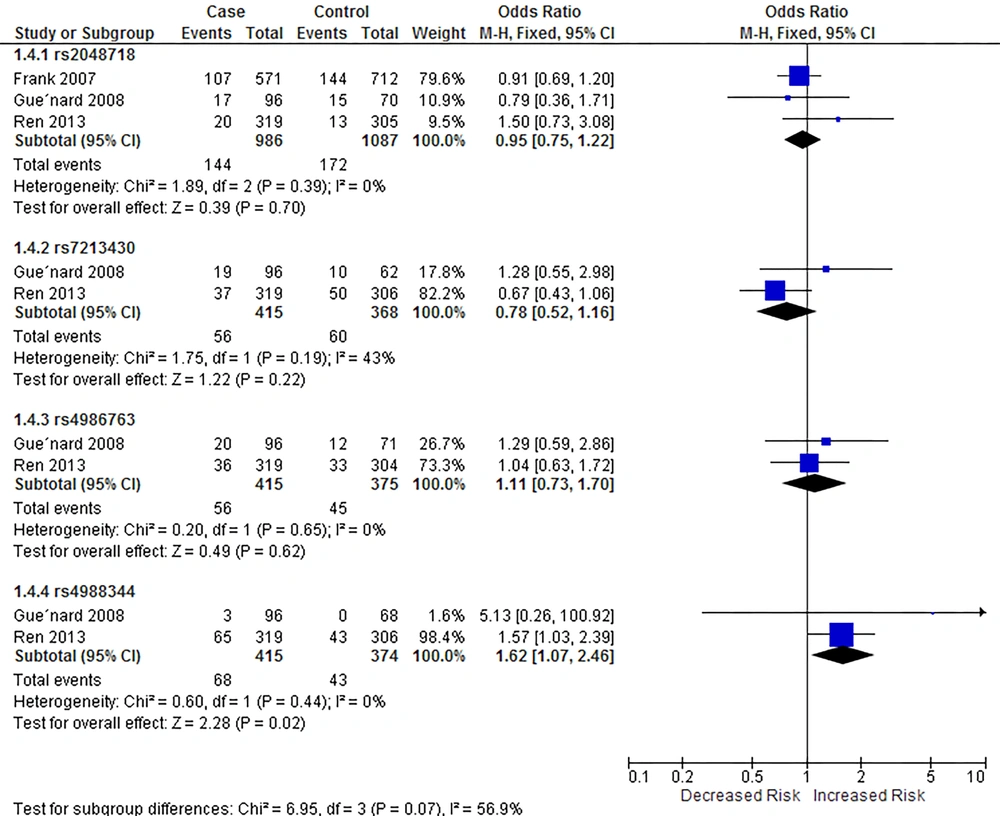
Our meta-analysis showed that rs4988344 was associated with breast cancer risk in homozygous (P = 0.46 for heterogeneity, OR = 1.66, 95% CI = [1.05, 2.63]) and recessive models (P = 0.44, OR = 1.62, 95% CI = [1.07, 2.46]), while rs7213430 was associated with breast cancer in dominant model (P = 0.03 for heterogeneity, OR = 0.74, 95% CI = [0.55, 0.98]). Figures 3 - 5 show the outcomes of the meta-analysis of associations between BRIP1 SNPs and breast cancer risk in homozygous, dominant, and recessive models, respectively.
Moreover, the funnel plots were illustrated to appraise the presence of publication bias in all assumed genetic models (Figure 6A-D). The overall results of the funnel plots demonstrated relatively symmetrical profiles indicative of low probability of publication bias in allelic and dominant models.
5. Discussion
BRIP1 has a direct interaction with the BRCT motif containing domain of BRCA1 and collaborates with BRCA1 in maintenance of genome instability and tumor suppressor effects (9). Moreover, inactivating truncating mutations in this gene are associated with both Fanconi anemia and vulnerability to breast cancer in homozygous and heterozygous states, respectively (10). Based on the emerging evidences regarding the role of BRCA1and Fanconi anemia-related genes in breast cancer susceptibility (7), we performed the present meta-analysis to evaluate the effects of SNPs within BRIP1 gene in conferring risk of breast cancer. We showed that rs4988344 was associated with breast cancer risk in homozygous and recessive models, while rs7213430 was associated with breast cancer in dominant model. The rs4988344 and rs7213430 have been located in 3’ untranslated region (3’UTR) and intron 5 of BRIP1, respectively (8). Ren et al. have reported the association between the A allele of rs7213430 and breast cancer risk Chinese Han population (8). The rs7213430 is located in the miR-101 seed-binding region and the sequence having G allele has a greater binding affinity with miR-101 than the sequence containing the A allele (11). Although the role of miR-101 as a tumour suppressor has been well documented in breast cancer (12), there are no data regarding its interaction with BRCA1 or BRIP1 in breast tissue. Future studies are needed to elaborate such possible interactions and their role in the pathogenesis of this kind of malignancy. The association between rs4988344 and breast cancer has been evaluated in Chinese Han population (8) as well as French Canadian individuals (7). Both studies failed to detect any significant association. However, the pooled data of these studies confirmed the association of this variant with breast cancer risk.
Our meta-analysis had some limitations. Firstly, the sample size was still small even after data pooling, which may influence the study power to appraise the association between the BRIP1 polymorphisms and predisposition to breast cancer. Secondly, we could not analyze the effects of environmental risk factors, such as menarche and menopause age, history of breastfeeding, and use of any hormonal medications, which might contribute in breast cancer risk. Based on the absence of such data, we could not assess gene-environment interactions in the present study. Moreover, based on the scarcity of data regarding the association between BRIP1 variants and breast cancer, we could not perform subgroup analysis. However, Begg’s test demonstrated low probability of publication bias, implying that the favored inclusion of positive results does not happen.
6. Conclusions
Our meta-analysis suggests that the rs4988344 within the BRIP1 gene was associated with breast cancer risk in homozygous and recessive models, while rs7213430 was associated with breast cancer in dominant model.
| Authors | Time | Region | Genetic Material | Ethnicity | Number of Cases | Number of Controls | Genotype Number in Controls | Genotype Number In Patients | NOS Score | Hardy-Weinberg Equilibrium/chi-Square | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WW | WM | MM | WW | WM | MM | |||||||||
| rs2048718 | ||||||||||||||
| Frank et al. (6) | 2007 | Germany | Blood | European | 571 | 712 | 228 | 340 | 144 | 181 | 283 | 107 | 7 | 0.4012/0.70 |
| Guénard et al. (7) | 2008 | Canada | Blood | French Canadian | 96 | 70 | 23 | 32 | 15 | 38 | 41 | 17 | 8 | 0.5379/0.38 |
| Ren et al. (8) | 2013 | China | Blood | Asian | 319 | 305 | 177 | 115 | 13 | 201 | 98 | 20 | 7 | 0.2883/1.13 |
| rs7213430 | ||||||||||||||
| Guénard et al. (7) | 2008 | Canada | Blood | French Canadian | 96 | 62 | 23 | 29 | 10 | 28 | 49 | 19 | 8 | 0.8656/0.03 |
| Ren et al. (8) | 2013 | China | Blood | Asian | 319 | 306 | 124 | 132 | 50 | 165 | 117 | 37 | 7 | 0.1433/2.14 |
| rs4986763 | ||||||||||||||
| Guénard et al. (7) | 2008 | Canada | Blood | French Canadian | 96 | 71 | 25 | 34 | 12 | 27 | 49 | 20 | 8 | 0.9393/0.006 |
| Ren et al. (8) | 2013 | China | Blood | Asian | 319 | 304 | 137 | 134 | 33 | 168 | 115 | 36 | 7 | 0.9782/0.000 |
| rs4988344 | ||||||||||||||
| Guénard et al. (7) | 2008 | Canada | Blood | French Canadian | 96 | 68 | 53 | 15 | 0 | 75 | 18 | 3 | 8 | 0.3066/1.04 |
| Ren et al. (8) | 2013 | China | Blood | Asian | 319 | 306 | 115 | 148 | 43 | 109 | 145 | 65 | 7 | 0.6744/0.18 |

![The funnel plots of the associations between BRIP1 SNPs and risk of breast cancer in all assumed inheritance models separately (Log [OR], logarithm of the odds ratio; perpendicular mark, mean effect size). The funnel plots of the associations between BRIP1 SNPs and risk of breast cancer in all assumed inheritance models separately (Log [OR], logarithm of the odds ratio; perpendicular mark, mean effect size).](https://services.brieflands.com/cdn/serve/3170b/1ca28ec877872c8efcde7b75f7dd2c12367889bf/ijcm-84234-i006-F6-preview.webp)