1. Background
The noxious effects of cancer chemotherapeutic agents on normal cells are one of the major limitations of using them. Thus, it is prominent to evaluate possible DNA damaging effects of anti-cancer drugs, which lead to probable secondary malignancies. DNA strand break is one of the biomarkers of genotoxicity (1).
Anthracyclines classify the most effective anti-cancer drugs ever developed and are widely used for chemotherapy of various types of cancers. Daunorubicin and doxorubicin were the first anthracyclines discovered, originally isolated from Streptomyces sp. The primary mechanism proposed for the anthracycline cytotoxicity is the inhibition of topoisomerase II. Other mechanisms of cytotoxicity include interference with helicase activity, intercalation between base pairs of the DNA/RNA strand, which lead to the inhibition of DNA and RNA synthesis, free radicals formation with consequent induction of DNA damage (2-4) or lipid peroxidation (5, 6). Finally, anthracyclines have been appeared to induce apoptotic cell death (7-9).
Idarubicin (4-demethoxy-daunorubicin), an anti-leukemic drug, is a member of anthracycline group used for chemotherapy of melanoma, sarcoma, lung, ovarian, and breast cancers (10, 11). Idarubicin was derived from daunorubicin as its synthetic analog after omitting a methoxy group that made it more lipophilic than daunorubicin and doxorubicin. Enhancing lipophilicity can increase its access to tumor cells and, therefore, raising its binding potential to DNA and genotoxicity. Binding and insertion to DNA prevent it from unwinding by interfering with the enzyme topoisomerase II (12-14). Some studies mentioned that idarubicin is 5 to 10 times more potent than daunorubicin and doxorubicin (15-17).
Dexrazoxane (ICRF-187) is a strong catalytic inhibitor of topoisomerase II, originally introduced as a chemotherapeutic agent, but it has just been found to exert cardioprotective effects against anthracyclines without reducing their antitumor efficacy or inducing new toxicities (18-21). This protection is due to its iron chelating properties. Also, dexrazoxane metabolites are able to remove iron from its complex with anthracyclines and, therefore, prevent the generation of reactive oxygen species (22-24). Dexrazoxane is useful clinically against accidental anthracycline extravasations (25).
Although dexrazoxane has been confirmed to have a protective effect against idarubicin-induced cardiotoxicity, its possible protective effects on genotoxicity of idarubicin have not been studied yet. The present research was undertaken to elucidate the potential genoprotective properties of dexrazoxane on oxidative DNA damage caused by idarubicin in HepG2 cells, using single cell gel electrophoresis or comet assay as a useful method for detecting DNA damage in individual cells (26). The most commonly used parameters in the comet assay method are tail moment, tail length, and percent of DNA in tail (27, 28). All these parameters were used in the present study to evaluate DNA damage. Lipid peroxidation was used as a marker of oxidative damage and a possible mechanism underlying this amelioration (29). We utilized thiobarbituric acid test for determination of lipid peroxidation extent, using thiobarbituric acid reactive substances (TBARS).
2. Methods
2.1. Chemicals
Idarubicin and dexrazoxane were respectively obtained fro Pharmacia (Italy) and sigma Co. (USA). Tris, Triton X-100, H2O2, NaCl, EDTA, NaOH, NaH2PO4, sodium dodecylsulfate, acetic acid, and n-butanol were procured from Merck Co. (Germany). Low melting point agarose (LMA), Na2HPO4, KCl, ethidium bromide, 2-thiobarbituric acid, pyridine, and 1,1,3,3-tetramethoxy propane were purchased from Sigma Co. (USA). Cinnagen Co. (Iran) provided normal melting point agarose (NMA). RPMI-1640, FBS, and antibiotics were supplied by PAA Co. (Australia). HepG2 cells were provided by Pasture institute (Iran).
2.2. Cell Culture
Human hepatocyte HepG2 cells were cultured in RPMI supplemented with 7% FBS and 1% penicillin/streptomycin. The cell culture was incubated at 37°C in humidified air (95%) and CO2 (5%) in micro-filter plates. The culture medium was renewed as needed, and when the cells reached 80 %, confluence was passaged.
2.3. Comet assay
2.3.1. Treatment
HepG2 cells were plated onto 25cm2 cell culture flask at a seeding density of 25 × 104 cells and allowed to adhere for 24 hours. To assess the suitable genotoxic concentration of idarubicin, cells were incubated with various concentrations (0.05, 0.5, 1, 5, and 10 µM) of idarubicin dissolved in culture medium for 1 hour. To investigate the protective effects of dexrazoxane, cells were exposed to its safe concentrations (10, 50, 100, and 200 µM) diluted in 1% DMSO in culture medium for 24 hours, followed by 1 hour exposure to genotoxic concentrations of idarubicin. After this period, washing cells was performed, using PBS and followed trypsinization for 5 minutes. After that and harvesting, the cell suspensions (1 × 104 cells/mL) were transferred to falcon tubes for the following steps.
To determine cell viability, cells were resuspended in PBS, mixed with trypan blue solution. Cell suspensions with viability > 80% were used.
2.3.2. Slide Preparation
Microscope slides were dipped into NMA (normal melting agarose) solution and they were dried. A mixture of 1 mL of LMA and 300 µL of above cell suspension were provided. This suspension was layered onto the slides and covered with a coverslip for 10 minutes 2 to 8°C. In lysis stage, we gently removed the coverslips and transferred slides in cold and freshly prepared lysing solution (pH = 10.0) under dark conditions for 40 minutes. The slides were, then, rinsed 3 times with deionized water to eliminate excess lysis solution. Next, for unwinding DNA, the slides were submerged in an alkaline solution (0.3 M NaOH, 1 mM EDTA, pH > 13) at room temperature for 40 minutes. Electrophoresis was performed in the new and same alkaline solution for 40 minutes at 25 V and 300 mA. After electrophoresis and in neutralization stage, we used 0.4 M Tris (pH 7.5), then, rinsed with water for 15 minutes and dried on a clean surface. Afterwards, the slides were stained with ethidium bromide (20 μg/mL) and rinsed with PBS and deionized water. All comet assay steps were carried out in dark conditions and all solutions were prepared fresh daily. Cells were examined under × 400 magnification, using fluorescence microscope. In the last stage, Comet score freeware (version 1.5) was used for control and treat analysis of comets in randomly selected cells (at least 100 cell per sample).
2.4. Measurement of Lipid Peroxidation
The extents of lipid peroxidation were determined by measuring TBARS levels in HepG2 cells according to Ohkawa et al. (30). Briefly, the cells were exposed to chosen concentrations of idarubicin and/or dexrazoxane. The cells were washed with PBS, scraped in 500 μL lysis solution on ice, and incubated for 40 minutes until efficient lysis was confirmed. The samples were, then, centrifuged at 13000 g for 2 minutes. Then, 200 µL of lysate supernatant, 200µL of 8.1% sodium dodecylsulfate, 1500 µL of acetic acid (pH 3.5), 1500 µL of 0.8% 2-thiobarbituricacid, and 600 µL of deionized water were mixed. The samples were, then, vortexed and heated, using a boiling water bath for 40 minutes and, then, cooled for 10 minutes. One mL of deionized water and 5 mL of n-butanol-pyridine solution were used to extract TBARS before centrifugation at 4000 rpm for 10 minutes. The organic layer was taken and the fluorescence was measured at emission and excitation wavelengths of 553 and 515 nm (synergy H1 multi-mode reader, USA). TBARS standards were prepared, using 1,1,3,3-tetramethoxypropane diluted in 40% ethanol to achieve concentrations of 2, 4, 6, 8, 10, and 0 µM. The levels of TBARS are expressed as nmol/mg of protein. The modified method of Lowry was employed to measure the protein concentration (31).
2.5. Statistical Analysis
The parameters used for the statistical evaluation of DNA damage were the tail moment, tail length, and percent of DNA in tail. The statistical significant differences were assessed by means of one-way analysis of variance (ANOVA), followed by suitable multiple comparison post hoc test. The differences were accepted to be significant if P < 0.05. Data on lipid peroxidation also analyzed, using ANOVA followed by Tukey’s multiple comparison.
3. Results
3.1. Genotoxic Effects of Idarubicin
The different concentrations of idarubicin (0.05, 0.5, 1, 5, and 10 µM) were incubated with HepG2 cells for 1 hour and the results were compared with negative control (cells incubated with RPMI) group. Among several parameters of genotoxicity, we used 3 common markers including tail length, % of DNA in tail, and tail moment for examination. Comet assay results revealed a significant difference (P < 0.001) between all tested concentrations and negative control for all above-mentioned parameters. The results are shown in Table 1.
aComparison of tail length, % DNA in tail and tail moment of HepG2 cells treated with different concentrations of idarubicin. Each data has been represented as Mean ± SEM.
bSignificant result (P < 0.0001) in compare with the control group.
3.2. Genoprotective Effects of Dexrazoxane
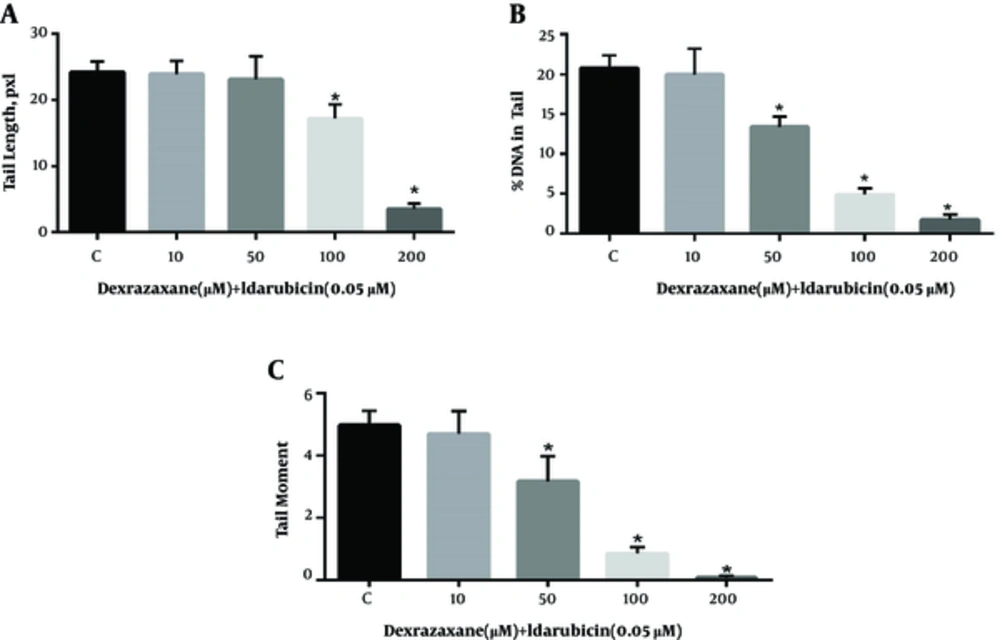
The results of comet assay after pre-treatment with dexrazoxane and exposure to idarubicin are shown in Figure 1. Treatment of cells to dexrazoxane alone did not result in any significant difference in the level of DNA strand breaks until 200 µM compared to negative control (data not shown). Thus, concentrations equal or less than 200 µM were supposed to be safe to be used in examination of genoprotective properties. Cells pre-treated with dexrazoxane following incubation with idarubicin showed significant decrease in the level of all parameters of DNA damage compared to idarubicin alone (control group).
Genoprotective effects of dexrazoxane on idarubicin induced DNA damage. Results of the comet assay performed on HepG2 cells pre-treated for 24-h with dexrazoxane followed by incubation for 1 hour with idarubicin. A, Tail length; B, % DNA in tail and C, Tail moment. Data are presented as Mean ± SEM of three replicates. The sign (*) shows significantly decreased results (P < 0.0001) in compare with the control (idarubicin alone) group.
3.3. Effect of Dexrazoxane on Idarubicin-Induced Lipid Peroxidation
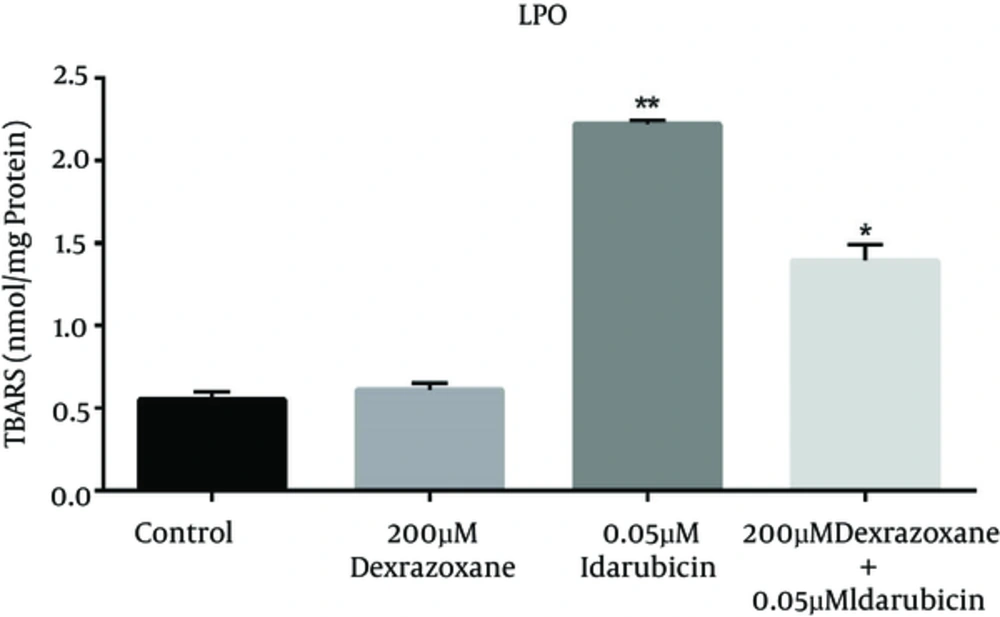
The effect of dexrazoxane on the idarubicin-induced lipid peroxidation (LPO) was assessed by measuring TBARS level in HepG2 cells. As shown in Figure 2, incubation of HepG2 cells with dexrazoxane (Dex) did not show significant difference compared to control (HepG2 cells incubated with RPMI), whereas idarubicin (Ida) treated cells represented extensive enhancement of TBARSs levels. Exposure to dexrazoxane in pre-treatment conditions exhibited a valuable decline in TBARS levels as compared to the data obtained after treatment with idarubicin alone.
Effect of dexrazoxane on idarubicin induced lipid peroxidation. Effects of dexrazoxane on lipid peroxidation levels (TBARS) induced by idarubicin in HepG2 cells (mean ± SD). The signs (*) and (**) show significantly differences results (P < 0.001) in compare with idarubicin alone group and (P < 0.0001) in compare with control group respectively.
4. Discussion
Dexrazoxane is still the only clearly effective cardioprotective agent used to counteract anthracyclines-induced cardiotoxicity. Therefore, we decided to evaluate possible genoprotective effects of dexrazoxane against DNA damage of idarubicin as a new anthracycline compound in order to reduce the unwanted toxicity in normal cells. This combination will provide more assurance on safe usage of increased drug doses in chemotherapy.
Idarubicin is an antibiotic chemotherapeutic agent, which is used in hematological malignancies. The superior DNA-binding capacity of idarubicin due to its higher lipophilicity leads to greater cytotoxicity compared to other anthracyclines. The first aim of this study was trying to find the lowest genotoxic concentration of idarubicin in cultured HepG2 cells by alkaline comet assay technique, a standard method for determining DNA damages including single- and double-strand DNA breaks (32). After comparison of different concentrations for all above parameters of DNA damage versus negative control group, we selected 0.05 µM of idarubicin as at least and optimum genotoxic concentration. Anthracyclines including idarubicin have quinone structure, permitting them to participate in electron transfer reactions mediated by oxoreductive enzymes. The reception of free electron converts it to semi-quinone free radicals and generation of reactive oxygen species (ROS), which may result in their DNA damage (8, 33).
Reactive oxygen species production by the complex metabolism of idarubicin could create abasic sites, and also induce single and double strand breaks. In fact, idarubicin, as topoisomerase II inhibitor drug, interacts with DNA and leads to cell death in higher doses (34).
This is in agreement with other studies, which represent a linear correlation between DNA bound anthracycline, DNA double strand breaks, and cell death. Although this effect is favorable in cancerous cells, DNA damage in normal cells increases the risk of secondary malignancies (35, 36). Thus, in order to increase safety and effectiveness of idarubicin, we attempted to evaluate the protective potential of dexrazoxane against DNA damage evoked by it.
The possible protective effects of dexrazoxane against the DNA damage of several genotoxic drugs were investigated (29, 37, 38). Furthermore, dexrazoxane has been reported to reduce ROS generation, lipid peroxidation, and oxidized glutathione (GSSG) accumulation (37). There are several reports documenting that dexrazoxane have inherent anti-oxidant activity, and ability to reduce the epirubicin-induced free radical production (24). Combination use of dexrazoxane did not disturb doxorubicin‘s distribution, metabolism or excretion; and indeed, the pharmacokinetics of anthracyclines remain unchanged (39).
The results of this study demonstrated that the treatment of HepG2 cells with dexrazoxane, 24 hours before idarubicin exposure, caused a noticeable decrease in DNA damage in comparison to idarubicin alone. Anti-genotoxic effects of dexrazoxane against doxorubicin in mouse ovarian cells were also reported (40). Their results revealed the ability of dexrazoxane to inhibit the topoisomerase II catalytic activity, to reduce double-strand DNA breaks and, thus, to prevent genotoxicity. Moreover, they found that dexrazoxane can protect against oxidative stress-induced DNA damage in a dose-dependent manner. They reported a concentration range of 20 to 200 µM of dexrazoxane in ameliorating effects that was similar to our protective concentration of dexrazoxane (100 and 200 µM).
Oxidative stress and production of ROS can result in DNA damage and degradation of protein and lipids, and is mainly accepted as one of the most important risk factors in the development of chronic diseases (41). Another result of free radical generation is lipid peroxidation, which is believed to be one of the causes of cardiovascular disease and cancer (42). The lipid peroxidation products mostly react with DNA, showing both genotoxic and mutagenic action.
In the present study, lipid peroxidation was assessed as an oxidative stress marker. TBARS level was measured after the cells were treated with idarubicin, compared with pretreatment of cells with dexrazoxane and control.
Our results demonstrated that treated cells with idarubicin showed the increased levels of TBARS compared to control. This study indicates that lipid peroxidation resulting in oxidative stress may contribute to the genotoxicity of idarubicin. However, pre-treatment of cells with dexrazoxane significantly decreased the level of TBARS, which demonstrates that dexrazoxane is able to ameliorate the lipid peroxidation caused by idarubicin. The potential of dexrazoxane for reduction of lipid peroxidation is in accordance with other observations (29, 37, 38).
4.1. Conclusions
In conclusion, the results of this study indicate that dexrazoxane was effective for the prevention of idarubicin-induced lipid peroxidation and DNA damage in HepG2 cells. Thus, dexrazoxane is able to attenuate deleterious effects of idarubicin in normal cells of patients with cancer in addition to its clinical application to prevent both anthracyclines-induced cardiotoxicity and extravasation (10, 20, 25). Further investigations are needed to focus on in vivo protective effects of dexrazoxane against idarubicin.