1. Background
Enterococci represent a substantial part of the gut flora and can exist under severe environmental conditions. They are Gram-positive cocci, facultative anaerobes, implicated in hospital-acquired infections superseded only by staphylococci as a source of Gram-positive nosocomial infections (1). Although more than 50 species have been reported (2), Enterococcus faecium and Enterococcus faecalis are the most clinically important multidrug-resistant infectious pathogens worldwide (3).
Vancomycin-resistant Enterococcus (VRE) particularly poses a major challenge to healthcare practitioners as its management has been trying in the hospital setting. The VRE infections have been reported to escalate cost and mortality as opposed to vancomycin-sensitive strains. Vancomycin-resistance in enterococci involves the modification of the peptidoglycan synthesis pathway.
Vancomycin attaches to the D-ala-D-ala end of the pentapeptide precursor, impeding the synthesis of the cell wall by inhibiting the cross-link of peptidoglycan chains. The VRE alters pentapeptide precursors, substituting the terminal D-ala with D-lactate or rarely D-ser (4), which now bind glycopeptides with significantly reduced affinity than typical precursors. The altered D-alanyl-D-lactate form causes loss of one hydrogen-bonding interaction and an interaction lesser than for D-alanyl-D-alanine association between vancomycin and the peptide, thereby conferring high-level resistance (1); nevertheless, D-alanyl-D-serine variation results in a six-fold affinity loss between vancomycin and the peptide, probably from steric impediment (5), conferring low-level vancomycin resistance (1).
Nine forms of vancomycin-resistance genotypes are expressed by enterococci, namely vanA to vanE, vanG, vanL, vanM, and vanN (1, 4, 6), with vanA and vanB of utmost clinical importance (1). The vanA genotype, most common globally and linked with vancomycin-resistance in enterococci in the hospital environment (1) confers resistance to vancomycin and teicoplanin (7). Moreover, less frequently, the vanB gene, mostly noticed in E. faecium, exhibits resistance to vancomycin but susceptibility to teicoplanin (7).
Bacterial attachment to host tissues is a key phase in the instigation of any infection process. Enterococci species express diverse virulence factors, including enterococcal surface protein (esp), aggregation substance (agg and asa1), gelatinase (gelE), cytolysin (cylA), hyaluronidase (hyl), pilA, pilB, ecbA, scm, fms8, efaAfs, efaAfm (adhesin-like endocarditis antigen A encoded by E. faecalis and E. faecium, respectively), and sgrA genes, which augment colonization, subsequent binding, and invasion in the host (8-10). Therefore, enterococci have numerous possibilities as most of the virulence genes commonly harbored are associated with adherence.
2. Objectives
Within Nigeria, there are reports of VRE recovery from the clinical setting (11) and even from food samples (9). However, documentation on the virulence of these organisms remains inadequate in Nigeria. Therefore, this study was performed to evaluate virulence factors and detect the possible presence of virulence and resistance genes in VRE from clinical samples from southwest Nigeria.
3. Methods
3.1. VRE from Participants
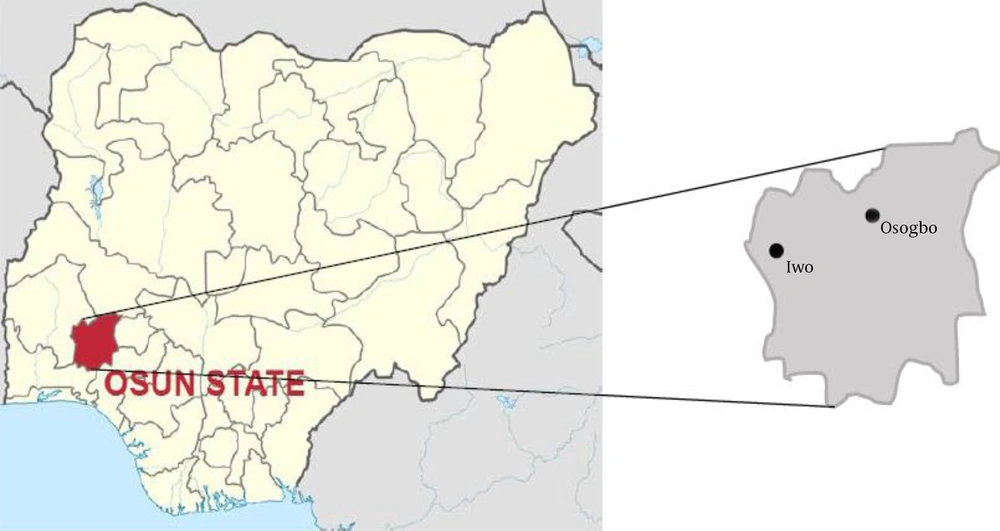
This study assessed 85 non-duplicate VRE isolates recovered from the samples in three selected hospitals in southwest Nigeria, namely State Specialist Hospital, Osogbo (7.76958°N; 4.54999°E), State Hospital, Iwo (7.66686°N; 4.19926°E), and Oke-Baale Primary Health Centre, Osogbo (7.76516°N; 4.578°E) (12) (Figure 1). Ethical approval was obtained from the Ethical Review Board of State Specialist Hospital, Osogbo (approval no.: HREC/SSHO/11/478).
The samples were aseptically inoculated into 10 mL of sterile Tryptone Soy Broth (TSB) (Oxoid, UK) and then streaked out on Slanetz and Bartley Agar. Pale-pink/maroon colonies on Slanetz and Bartley Agar indicates the growth of enterococci species. Overnight growth from Slanetz and Bartley Agar plates were inoculated onto Bile Aesculin Agar and Mannitol Salt Agar for the speciation of enterococci into E. faecalis (yellow colonies on Mannitol Salt Agar [MSA]) and E. faecium (no growth on MSA). Screening for vancomycin resistance was through inoculation on Brain Heart Infusion Agar supplemented with 6 µg/mL of vancomycin. Recovered isolates were preserved in TSB supplemented with 15% glycerol, frozen, and stored at -20°C.
3.2. Screening for Virulence Determinants
3.2.1. Gelatinase Production
Overnight cultures of VRE isolates were stabbed 4 - 5 times 1/2 inch depth into freshly prepared nutrient-gelatin medium and incubated at 37 ± 2°C for 48 h, along with an un-inoculated tube. Afterward, the tubes were removed without shaking or inversion, refrigerated, then gently inverted, and visually observed for gelatinase production as indicated by partial or complete liquefaction of the test media at 4°C. Control and gelatinase negative tubes remained solid.
3.2.2. Hemolysin/Cytolysin Production
The VRE isolates were streaked on freshly-prepared blood agar plates, incubated for 24 - 48 h at 37 ± 2°C, and visually observed for the patterns of hemolysis. Hemolysin production was scored as β (complete), α (partial), or γ (no clear zone) hemolysis indicated by clear/colorless zone, greenish zone, and completes absence of hemolysis, respectively.
3.2.3. Biofilm Production
The test isolate was inoculated on Congo Red Agar. The production of black colonies with a dry crystalline consistency indicated biofilm production; nonetheless, non-biofilm producers developed red colonies.
3.2.4. DNase Production
Overnight cultures of test isolates were inoculated onto DNase Agar and then incubated at 37 ± 2°C for 18 - 24 h. The plates were then flooded with 1N HCl for a few minutes, excess HCl tipped off, and the plates were observed within 5 min against a dark background for clear zones surrounding the line of the streak, indicative of DNase production.
3.2.5. Caseinase Production
Test isolates were inoculated onto Mueller-Hinton Agar supplemented with 3% skimmed milk and then incubated at 37 ± 2°C overnight. The development of clear proteolytic zones around the line of the streak was indicative of caseinase production.
3.3. Molecular Characterization of Selected Bacterial Isolates
3.3.1. Extraction of Chromosomal DNA
Deoxyribonucleic acid (DNA) was extracted from VRE isolates by thermal lysis of the cell. A 1 mL aliquot of an overnight culture of VRE was centrifuged at 5,000 rpm for 10 min, and the supernatants were discarded. The pellets were re-suspended in 500 μL of sterile DNA/DNase/RNase free water, vortexed, centrifuged at 5,000 rpm for 1 min, and then re-suspended in 200 μL of sterile water. The suspension was boiled in a water bath at 100°C for 10 min with lids closed, lysates cooled to room temperature, and then centrifuged at 10,000 rpm for 10 min. The purity and concentration of the extracted DNA in the supernatant were estimated using NanoDrop One spectrophotometer (Thermo Fisher Scientific, United States) at 260 nm and stored at -20°C in sterile Eppendorf tubes to serve as DNA templates for subsequent molecular characterization.
3.3.2. Identification of Bacterial Isolates and Detection of Target Genes
The DNA templates were subjected to a polymerase chain reaction (PCR) using species-specific primers for ddlE genes (Inqaba Biotec, South Africa) to confirm the biochemical identification of the vancomycin-resistant E. faecium and E. faecalis isolates (13). Virulence genes were detected using four multiplex PCR reactions only in isolates confirmed genotypically as VRE faecium and faecalis. Virulence genes, namely asa1, gelE, cylA, esp, and hyl, were amplified in one reaction (14); however, pilA-pilB-efaAfm, fms8-sgrA, and ecbA-scm were run in three other reactions. The resistance genes vanA and vanB were also screened for using Multiplex PCR (Table 1). Amplification was carried out using the Master Cycler Nexus Gradient 230 (Eppendorf, Germany) (Table 2) in a total volume of 25 μL solution containing 12.5 μL of 2× Master-Mix (Biolabs, England), 0.5 μL of 10 µM each of forward and reverse primers (Inqaba Biotec, South Africa), and 5 μL of each DNA template made up to 25 μL with DNAse/RNAse free sterile water (Bio-Concept, New Hampshire, United States).
| Target Genes Primers | Sequence (5’ - 3’) | Band Size (bp) | References |
|---|---|---|---|
| Identification: E. faecium and E. faecalis | |||
| ddlE faecium | 658 | (11) | |
| F | TTGAGGCAGACCAGATTGACG | ||
| R | TATGACAGCGACTCCGATTCC | ||
| ddlE faecalis | 941 | (11) | |
| F | ATCAAGTACAGTTAGTCTTTATTAG | ||
| R | ACGATTCAAAGCTAACTGAATCAGT | ||
| Vancomycin-Resistant Gene | |||
| van A | 732 | (11) | |
| F | GGGAAAACGACAATTGCC | ||
| R | GTACAATGCGGCCGTTA | ||
| van B | 635 | (11) | |
| F | ATGGGAAGCCGATAGTC | ||
| R | GATTTCGTTCCTCGACC | ||
| Virulence Genes | |||
| asa1 | 375 | (12) | |
| ASA11 | GCACGCTATTACGAACTATGA | ||
| ASA12 | TAAGAAAGAACATCACCACGA | ||
| gelE | 213 | (12) | |
| GEL11 | TATGACAATGCTTTTTGGGAT | ||
| GEL12 | AGATGCACCCGAAATAATATA | ||
| cylA | 688 | (12) | |
| CYT I | ACTCGGGGATTGATAGGC | ||
| CYT IIb | GCTGCTAAAGCTGCGCTT | ||
| esp | 510 | (12) | |
| ESP14F | AGATTTCATCTTTGATTCTTGG | ||
| ESP12R | AATTGATTCTTTAGCATCTGG | ||
| hyl | 276 | (12) | |
| HYL1 | ACAGAAGAGCTGCAGGAAATG | ||
| HYL2 | GACTGACGTCCAAGTTTCCAA | ||
| pilA | 459 | (6) | |
| F | AAAACGCCACCAGAGAAGGT | ||
| R | CATTGGCGCAATCACAACCA | ||
| pilB | 959 | (6) | |
| F | GATACCCAGCTGACGGCTTT | ||
| R | GGTACTGCCGAAAACGAAGC | ||
| fms8 | 765 | (6) | |
| F | AGACGAGCAGATGAACAGCC | ||
| R | CCCGTCAATCGTCGTACTGT | ||
| EfaAfm | 199 | (6) | |
| F | AAAAGGCAAGCGACGCAGAT | ||
| R | AGGTCTAGCCAAGCATGAGG | ||
| sgrA | 150 | (6) | |
| F | CTGATCGGATTGTTTATGA | ||
| R | AATAAACTTCCCCAATAACTT | ||
| ecbA | 182 | (6) | |
| F | GGAGTGAGGCTTTTAAACCAGA | ||
| R | GGAAACAGGGTACTTTG | ||
| scm | 1015 | (6) | |
| F | GTTTACTAGTCCTAGTTGC | ||
| R | TCTGTACTGTCGCTTGTGTC | ||
Primer Sequences for Vancomycin-Resistant Enterococci Identification and Characterization
| Target Gene | Initial Denaturation | No. of Cycles | Temperature (°C), Time | Final Extension | ||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||
| ddlE faecium; ddlE faecalis | 95°C, 5 min | 30 | 95°C, 30 s | 53°C, 30 s | 72°C, 60 s | 72°C, 10 min |
| vanA-vanB | 94°C, 3 min | 30 | 94°C, 60 s | 54°C, 60 s | 72°C, 60 s | 72°C, 7 min |
| asa1-gelE-cylA-esp-hyl | 95°C, 15 min | 30 | 94°C, 60 s | 56°C, 60 s | 72°C, 60 s | 72°C, 10 min |
| pilA-pilB-efaAfm | 95°C, 2 min | 35 | 95°C, 20 s | 58°C, 10 s | 72°C, 20 s | 72°C, 5 min |
| fms8-sgrA | 95°C, 2 min | 35 | 95°C, 20 s | 58°C, 10 s | 72°C, 20 s | 72°C, 5 min |
| ecbA-scm | 95°C, 2 min | 35 | 95°C, 20 s | 58°C, 10 s | 72°C, 20 s | 72°C, 5 min |
Polymerase Chain Reaction Protocols for Molecular Characterization of Vancomycin-Resistant Enterococci
Each amplicon (10 μL) was run on 1.0% agarose gel stained with SafeView Classic at 80 V for 60 min for isolate identification and resistance gene detection and at 100 V for 60 min for detection of virulence genes. The gels were visualized with the UV trans-illuminator E-BOX-CX5 TS imaging system (Vilber, France). Moreover, the 100 bp DNA Ladder (Biolabs, England) served as DNA molecular weight standard. E. faecalis ATCC 51299 and E. faecalis MMH594 were the positive control strains.
3.3.3. Plasmid Profile Analyses
Plasmid DNA from VRE isolates was extracted using Plasmid Miniprep Kit (ZymoPURE, California), and the eluted plasmid DNA was stored at ≤ -20°C. The vanA and vanB genes were screened with Multiplex PCR in a total volume of 25 μL reaction and observed as described above.
4. Results
4.1. Screening for Virulence Determinants
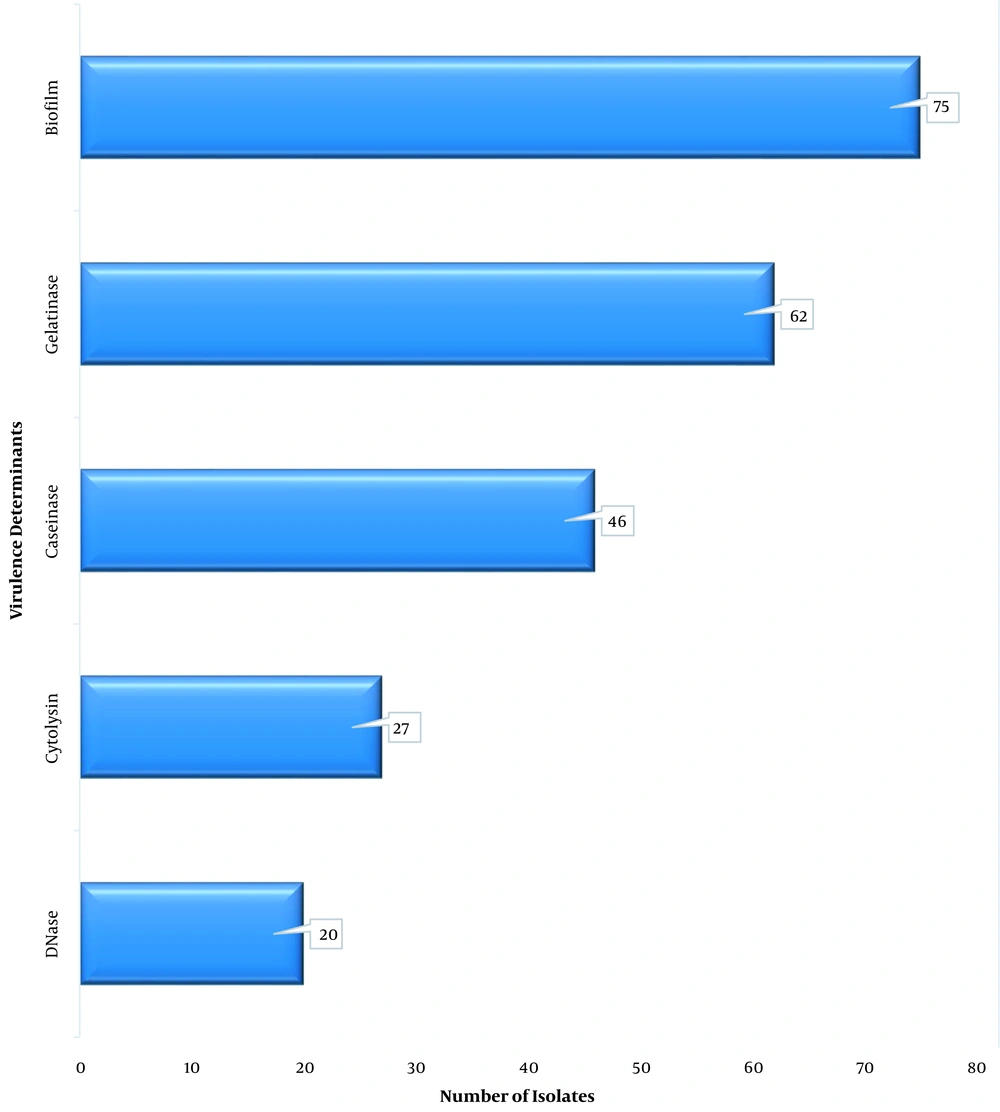
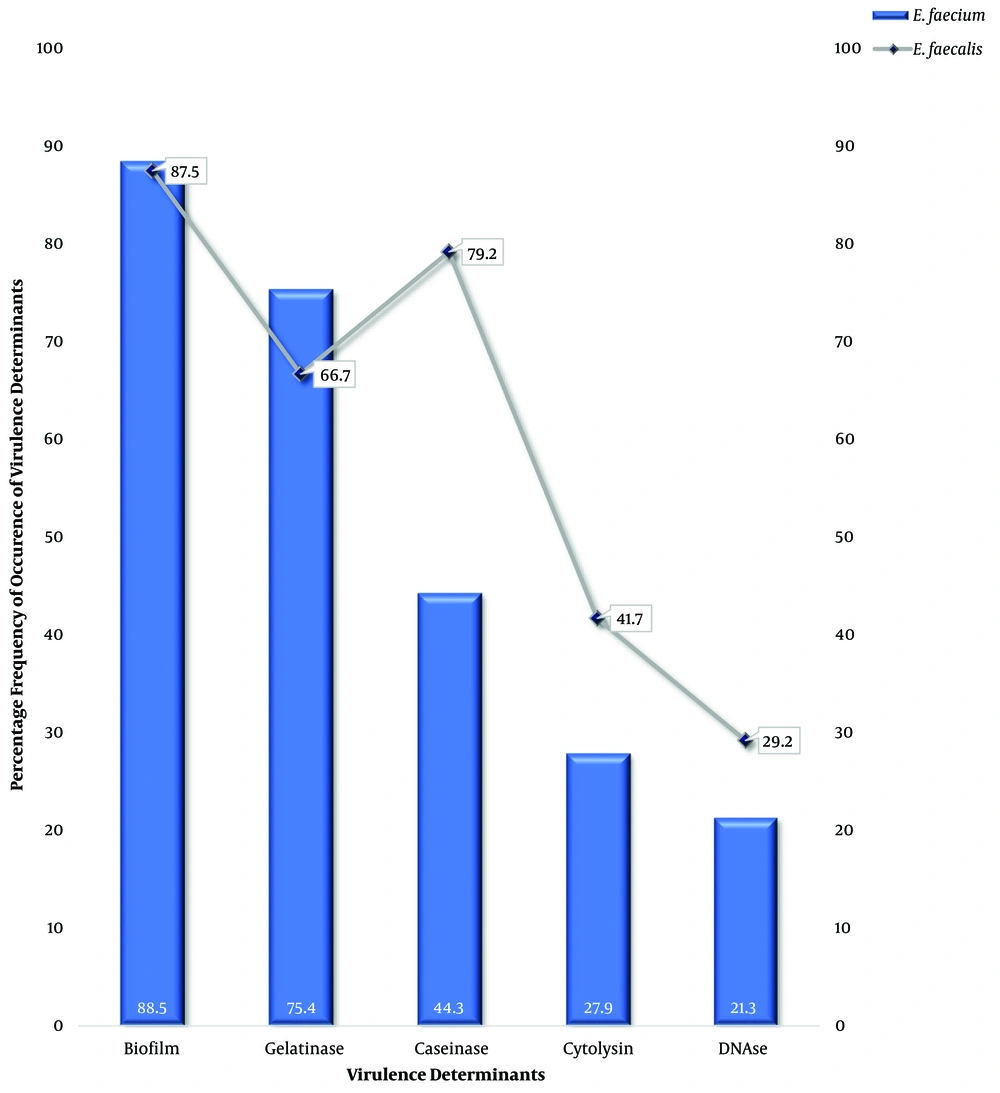
All 85 VRE isolates (100%) were observed to be catalase-negative, and all grew in the presence of 40% bile; however, 84 VRE isolates (98.8%) hydrolyzed aesculin. Only 27 VRE isolates (31.8%) could produce cytolysin on blood agar. The biofilm producers were 75 VRE isolates (88.2%). Furthermore, 72.9% of VRE isolates produced a gelatinase enzyme; nevertheless, only 20 VRE isolates (23.5%) produced DNase (Figure 2). Six isolates (7.0%) produced all five virulence factors. Only two isolates (2.4%) produced no virulence factor, as all the others produced two virulence factors or more. Biofilm was the most expressed factor at 88.5% and 87.5% for E. faecium and E. faecalis, respectively; nonetheless, the least virulence factor was DNase at 21.3% and 29.2% for E. faecium and E. faecalis, respectively (Figure 3).
4.2. Molecular Characterization
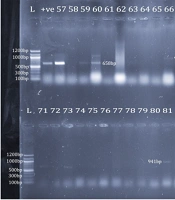
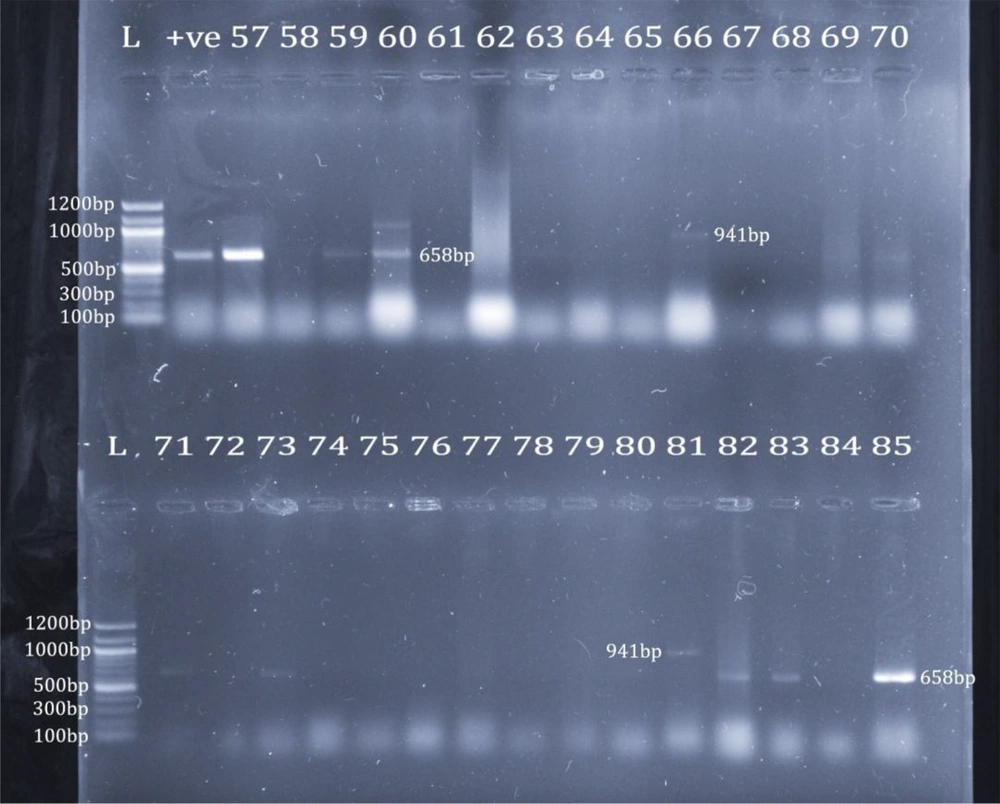
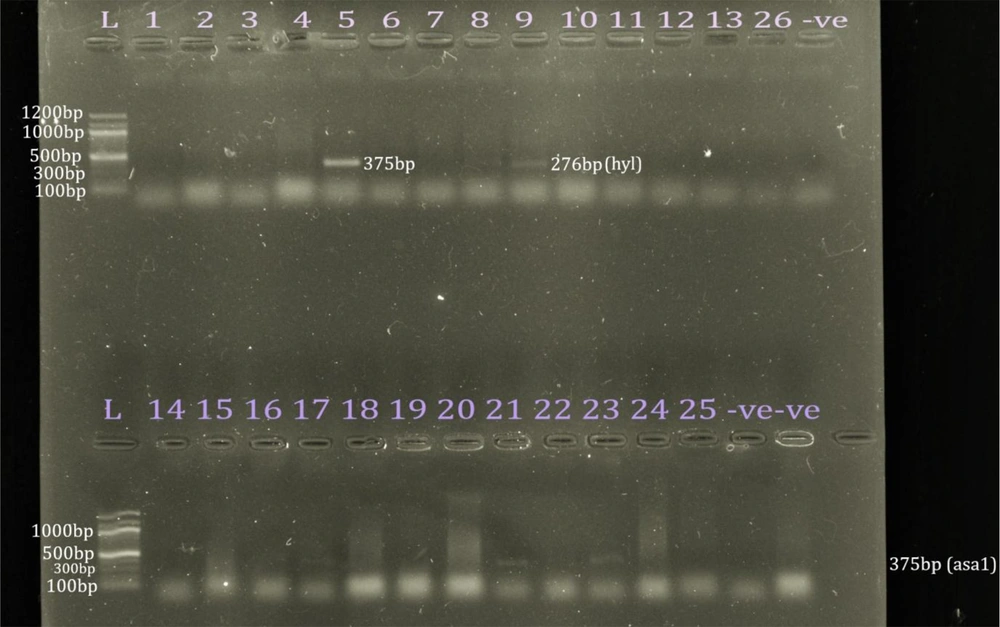
Only 25 isolates (29.4%) revealed bands for the identification with ddlE primers. In addition, 23 isolates (92.0%) were E. faecium; however, two isolates (8.0%) were E. faecalis. Using phenotypic identification, 23 out of 61 isolates phenotypically identified as E. faecium (37.7%) revealed bands for E. faecium identification; nevertheless, only 2 out of 24 isolates phenotypically identified as E. faecalis (8.3%) revealed bands for E. faecalis identification (Figure 4). The 25 VRE isolates confirmed genotypically as E. faecium and E. faecalis were selected for screening for virulence genes. Five isolates (20%) revealed bands, including four isolates for asa1 (16%) and only one isolate for hyl (4%). None of the other 10 virulence genes were detected (Figure 5). Only one isolate showed a band for the vanB gene at 635 bp; none of them had the vanA gene.
Identification of E. faecium and E. faecalis. Upper row: lane 1: 100 bp marker; lane 2: ddlE. faecium positive control; lanes 3, 5, and 6: E. faecium (658 bp); lane 12: E. faecalis (941 bp); lower row: lane 1: 100 bp marker; lanes 2, 4, 13, 14, and 16: E. faecium (658 bp); lane 12: E. faecalis (941 bp)
5. Discussion
The virulence factors of enterococci play a major role in the pathogenicity of enterococci and could be explained not only by the presence of virulence determinants; antibiotic resistance genes play an important role in the pathogenicity of enterococcal strains (15, 16). In-vivo and on medical devices biofilm formation aids disease development as it boosts the persistence of infections and reduces antimicrobial activity (17). Biofilm production was 88.5% and 87.5% in E. faecium and E. faecalis, respectively, corroborating an earlier study (18) where biofilm production was 86.6% among enterococci isolates. GelE, a foremost virulence determinant among biofilm producers (18), facilitates signals within the quorum sensing fsr system resulting in biofilm production (16); however, earlier studies postulate that no correlation is observed between gelatinase and biofilm production in many E. faecalis isolates (19).
In this study, gelatinase production was higher in E. faecium than E. faecalis, a finding at variance with another report (20) with lower rates for both species but higher production by E. faecalis than E. faecium. Numerous E. faecalis isolates in the current study (64.7%; 55/85) coproduced biofilm and gelatinase. This may be adduced to environmental and genetic factors, virulence, and the existence of other mechanisms as these affect surface activity and intercellular interactions (10).
A study reported no production of gelatinase in some Enterococcus isolates, although gelE was detected (21). The activation of gelE expression has been reported in the late exponential growth phase at high cell concentrations, and its intracellular expression can raise the severity of infections. Biofilm formation is independent of the presence or lack of the esp gene (16, 18, 22); nevertheless, other authors affirmed the positive relationship between the presence of esp (23) and asa1 gene with biofilm formation in enterococci as asa1 gene promotes the adherence of microorganisms to surfaces (16, 18). However, no gelE or esp gene was detected in all the isolates of the present study, and only 4 out of 25 screened isolates had the asa1 gene, reinforcing the complexity of the processes involved in Enterococcus virulence.
Cytolysin facilitates infection by damaging cell membranes (16, 20) and has been reported to enhance virulence in animal models (16). Hemolysin and/or gelatinase aids nutrient acquisition from host tissues and advances invasion, thereby increasing the severity of human infections. However, the failure to detect the cylA gene in the isolates of the present study, in line with other studies (18), underscores the need for phenotypic and molecular screening for virulence.
DNase production in this study was low for E. faecium (21.3%) and E. faecalis (29.2%), respectively. DNase hydrolyzes nucleic acids, contributing to bacterial virulence, although E. faecium is reported to be devoid of DNase activity (24). Hyaluronidase, which was detected in only 4.0% of the isolates of this study, is encoded by chromosomal hyl and degrades hyaluronate. Bacterial hyaluronidase behaves as endo-N-acetylhexosaminidase, destroys β-1-4 linkage, consequently creating unsaturated disaccharides, causing tissue damage (16).
Pathogenicity is related to the ability of virulent strains to grow profusely in the intestinal tract and invade the body. Host factors, such as underlying medical conditions, immune status, and antibiotics exposure, are thought to play a role in the pathogenicity of enterococci. Low recovery of virulence genes in this study population suggests strongly that virulence alone might not indicate infection, as other mediators of pathogenicity could be left unexplained (25).
The present study detected the vanB gene in only one isolate, but vanA in none. This result is substantiated by a study where multi-resistance E. faecium strains had no vancomycin-resistance genes, E. faecalis strains ST774 carried the vanB gene, and ST133 had no acquired resistance genes as confirmed by vancomycin susceptibility testing (26), a finding at variance with other reports (27). In another study, isolates screened as vanA and vanB phenotypes were negative for both genes; nonetheless, they were positive for a fragment of the vanHM gene (28). Therefore, the results of the current study suggest the presence of other van genotypes the detection of which might be missed (28). However, a major limitation of this study was the inability to confirm the identification using molecular methods and to screen all the enterococcal isolates for other virulence genes due to limited resources.