1. Background
Staphylococcus aureus is the most infectious Staphylococcus species and the most common bacterium isolated from hospital-acquired infections (1). It is also the second most prevalent pathogen among patients in outpatient clinics. S. aureus can cause mild to severe infections such as skin and soft tissue infections (SSTIs) and sepsis (1). Additionally, it may lead to toxin-mediated infections that are associated with high mortality. Within a decade of discovery of penicillin, about 90% of the S. aureus strains isolated from hospitals became resistant to penicillin by producing beta-lactamases. Hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA) includes the strains that are resistant to all b-lactams including penicillins and cephalosporins and are isolated in healthcare environments (2). MSRA rates increased steadily until the late 1990s when it experienced a sharp increase in rate (2). This was followed by the appearance of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) when community-acquired infections were identified with MRSA in healthy people with limited or no exposure to predisposing factors of MRSA (1). The mecA gene is responsible for methicillin resistance, which is carried by cassette chromosome mec (SCCmec) (3).
Life-threatening infections are predominantly associated with CA-MRSA that are more virulent due to carrying Panton–Valentine Leukocidin (PVL). PVL had been found mainly within CA-MSSA infections even before the emergence of MRSA. It is encoded by genes lukS-PV and lukF-PV that comprise two subunits that form pores in the cell wall of leukocytes by producing a substance called leucocidin (4).
In Iran, the prevalence of MRSA in healthcare workers was 16% - 35% (5, 6). The MRSA rate was 10.1% among healthcare workers from Jordan (7), 73% among healthcare workers from Saudi Arabia (8), and 22.5% in healthcare workers from Iraq (9). The prevalence of MRSA carriage was also studied in Iraq. The prevalence of MRSA was found to be 4.2% (10). In another study that was performed in urban and rural schools, the overall nasal carriage rates of S. aureus and MRSA isolates were 17.75% and 10%, respectively (11). MRSA infections are spreading steadily at hospitals and communities, making a public health problem. In order to control MRSA-related infections, the continuous monitoring of the prevalence of S. aureus is needed (1). Continuous study and monitoring of MRSA are critical for the prevention and treatment of MRSA-related infections (12). Such studies can provide information on the spread of the bacterium, the trends of infections and the appearance of new strains in hospitals and communities, the antibiogram and antibiotic sensitivity patterns, and the risk factors associated with infection (12). Such surveillance helps healthcare providers to plan prevention strategies and clinicians to provide accurate empiric antibiotics for infections circulating in their regions with a subsequent improvement of infection treatment outcomes (1).
2. Objectives
The aim of this study was to assess and compare the prevalence of CA-MRSA and HA-MRSA in Duhok city, Kurdistan region, Iraq, and detect mecA and PVL genes among two healthy groups of people, community students and hospital staff.
3. Methods
3.1. Sample Collection
With a confidence interval of 95% (Z = 1.96), an error margin of 10%, a design effect of 1, and an expected response rate of 0.8, the sample size of 91 was calculated. The samples were collected in the period between January 2018 and June 2018. Students who had a history of one of the following conditions were excluded: recent admission to healthcare facilities, recent surgical operations, end-stage kidney diseases on hemodialysis, using intravenous cannula at the time of swab taking, MRSA detection, and active skin infections. Both categories of samples were taken from nares by using sterile swabs, inoculated into transport media, and examined immediately after reaching the lab. All healthcare workers who agreed to participate and signed consent forms were included in the study. Healthcare workers who did not agree to participate were excluded.
3.2. Laboratory Investigation
3.2.1. Conventional Tests
All isolates were evaluated by several conventional laboratory tests for detecting and identifying S. aureus and MRSA among hospital staff and community students. These tests included Gram staining and biochemical tests such as catalase and tube coagulase tests. Furthermore, all samples were cultured on Mannitol Salt Agar (Oxoid) for fermentation and Brain Heart Infusion (BHI) (Oxoid). Moreover, Mueller-Hinton Agar (Oxoid Limited, Hampshire, England) was used for antibiotic sensitivity testing to oxacillin by using Kirby-Bauer disk diffusion as recommended by the CLIS guidelines (13).
3.2.2. Molecular Analyses
3.2.2.1. Detection of mecA and PVL Genes by Polymerase Chain Reaction (PCR)
PCR was applied to all S. aureus isolates for the detection of mecA and PVL genes according to instructions by Murakami et al. (14). The forward (GTG GAA TTG GCC AAT ACA GG) and reverse (TAG GTT CTG CAG TAC CGG AT) primers were used for the mecA gene. PCR amplification conditions for the mecA gene were 35 cycles starting with initial activation step for 2 min per each cycle at 95ºC, followed by denaturation step for 30 s at 95ºC. The annealing time was 1 min at 58ºC and extension was 2 min at 72ºC (15, 16). Luk-PV-1 (ATCATTAGGTAAAATGTCTGGACATGATCCA) and LukPV- (GCATCAAGTGTA TTGG ATAGCAAAAGC) primers were used for the PVL gene. The amplification conditions for PVL started with initial activation for 2 min at 95ºC, followed by denaturation for 30 s at 95ºC. Then, the annealing step was performed for 1 min at 55ºC, followed by primer extension for 2 min at 72ºC. The reaction was done for 35 thermal cycles (17).
3.3. Ethical Statement
Consent was taken from hospital staff and community students to use their samples for research purposes. The study was approved by the Scientific Committee of Directorate of Health, Duhok, Kurdistan region of Iraq.
4. Results
4.1. Staphylococcus aureus Isolates Among Hospital Staff
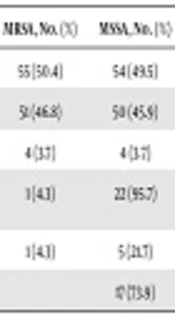
In the present study, 109 samples were collected from healthcare workers at different health facilities in Duhok city, Kurdistan region of Iraq. There were 67/109 (61.4%) males in the sample. The age range of healthcare workers was 18 - 49 years. Additionally, 103 samples were collected from secondary schools in the city; 78/103 (75.7%) samples were from males with the age range of 18 - 22 years. All the isolates from hospital staff were identified as S. aureus according to the standard identification procedure. Based on the genotypic analysis, 55 of 109 (50.4%) isolates were MRSA carrying the mecA gene and 4/55 (3.7%) were MRSA-bearing PVL genes. Additionally, 54/109 (49.5%) isolates were MSSA among which, 4/54 (3.7%) isolates were MSSA-PVL gene-positive (Table 1).
| Sample Categories | Total Samples | MRSA, No. (%) | MSSA, No. (%) |
|---|---|---|---|
| Hospital staff | 109 | 55 (50.4) | 54 (49.5) |
| PVL- | 51 (46.8) | 50 (45.9) | |
| PVL+ | 4 (3.7) | 4 (3.7) | |
| Community students | 103 | 1 (4.3) | 22 (95.7) |
| PVL+ | 1 (4.3) | 5 (21.7) | |
| PVL- | 17 (73.9) |
4.2. Staphylococcus aureus Isolates Among Community Students
The results showed that 23/103 (22.3%) samples from community students were determined as S. aureus. There were 22 MSSA strains among which, 5/22 (21.7%) isolates were MSSA carrying the PVL gene whereas 17/22 (73.9%) isolates were MSSA PVL gene-negative. Moreover, only was 1/23 (4.3%) isolate determined as MRSA that was carrying the PVL gene (Table 1).
5. Discussion
In the present study, all healthcare workers were found to be S. aureus carriers among whom, 50.4% of the isolates were identified as MRSA that carried the mecA gene. A comparable rate was found in a previous study carried out in Duhok where MRSA was found in 51.7% of the isolated S. aureus strains in healthcare workers, implying that MRSA is prevalent at hospitals in Kurdistan (15).
Based on the genotypic analysis, 55 out of 109 (50.4%) isolates were MRSA carrying the mecA gene. Among healthcare worker isolates, 7% carried the PVL gene including 3.7% MRSA-PVL-positive isolates and 3.7% MSSA-PVL gene-positive isolates. Noteworthy, these data were comparable with the results of a previous study that was performed in Duhok in 2016 among healthcare workers (15). A similar rate of PVL positivity was found in MRSA strains isolated from hospitals in the Netherland where about 8% of the isolates carried the PVL gene. In Iran, the prevalence of MRSA, the PVL gene, and MRSA-PVL isolates were 32%, 19%, and 10%, respectively (18). In Germany, the prevalence of MRSA isolates among healthcare workers was 11.3%, among which 9.1% of the isolates were positive for PVL (19).
On the other hand, about 22.3% of the nasal carriage isolates from community students were found to be S. aureus that was comparable to the rate found in northern Pakistan (24%) (20). In contrast, a higher rate was reported in the United State where 28.6% of the recruited samples carried S. aureus. In 2014, a study was conducted in Duhok city to evaluate the nasal carriage of CA-MRSA and the results showed that 18.4% and 2.04% carried S. aureus and MRSA, respectively (21). Additionally, a similar rate of MRSA was shown among university students with the rates of 17.5% and 4.2% for S. aureus and MRSA, respectively (10). Moreover, the MRSA rate among community students that carried the PVL gene in the current study was 4.3%, which is higher than the rate (2.5%) found among students in Palestine in 2011 (20) and likewise the rate (2.4%) in Brazil (22). Interestingly, the prevalence rates of HA-MRSA and CA-MRSA carrying PVL were 3.7% and 4.3%, respectively. More studies are needed to monitor the prevalence rates of PVL and its evolution over time.
To conclude, a higher prevalence of MRSA was found in healthcare workers whereas the MRSA isolates were carrying a closely similar rate of PVL gene in both groups. Moreover, CA-MSSA was carrying the highest rate of the PVL gene. Additionally, continuous surveillance is needed to reduce the spread of isolates between hospital staff carriers and patients, as well as in the community.