Introduction
Immune thrombocytopenic purpura (ITP) known as Idiopathic thrombocytopenic purpura is a multifactorial autoimmune bleeding disease associated with platelet destruction and discriminated by isolated thrombocytopenia (platelet count < 150,000 u/L) that was reported in almost 2 per 100,000 adults with a mean age of diagnosis of 50 years (1, 2). However, the vague pathogenesis, the abnormalities in the number and the function of different immune cells can play a crucial role in this disease. ITP phenotype, characterized by dysfunctional T-lymphocyte immunity, dysregulation in pre-B-cell, and T cell immunophenotypic markers, was recognized in bone marrow lymphocytes of pediatric ITP (3, 4). Besides, it is believed that membrane glycoproteins IIb-IIIa of platelet was targeted by immunoglobulin G autoantibody which is confirmed significantly by elevated CRP levels in ITP patients (5,6). These autoantibodies are recognized in 40–60% of patients and provide condition to Kupffer cells and splenic macrophages in the liver phagocytosis platelets (7). Other mechanisms include impaired production of platelet stimulatory hormone, thrombopoietin, reduced expression of human leukocyte antigen-G and immunoglobulin-like transcripts or secondary contributors such as childhood exposure to viruses, helicobacter pylori infection, and pregnancy (8-10). Zhang, et al., determined six marker proteins that separate primary ITP from secondary ITP, including NPS, EDN1, CORT, CLEC7A, CCL18, and NPPB. Most of the detected proteins related to the immune system act as up/down-regulator in macrophages and platelet (11). Platelets can be recognized with the expression of CD38 as a prognostic marker for ITP (2).
As mentioned before, ITP classified as acute and chronic types and sub-categorized by primary and secondary etiology (9, 10). Besides, the alternative classification by international consensus guidelines organized 3 phases as newly diagnosed (up to 3 months), persistent (3-12 months’ duration), and chronic (over 12 months’ duration) (2, 6 and 11).
ITP patients were characterized by a decrease in platelet count of peripheral blood and variable bleeding symptoms. In severe cases, it may lead to fatal intracranial hemorrhage. Thus, prompt diagnosis and early therapeutic intervention are essential (12, 13).
There are no specific criteria for diagnosing ITP, and diagnosis is based on the exclusion criteria of the other diseases, such as lupus erythematosus, Von Willebrand disease type IIb, hemolytic uremic syndrome, Evans syndrome, disseminated intravascular coagulation, Posttransfusion purpura, paroxysmal nocturnal hemoglobinuria, myelodysplastic syndrome, lymphoproliferative disorders, Infections (viral, bacterial, parasitic), and drug-induced thrombocytopenia. Furthermore, antiplatelet antibody testing is not recommended because of high inter-laboratory variability and reduced sensitivity (14-16).
Microarray technology is a prevalent technique for studying the pattern of expression of a large number of genes to analyze a genome. Microarray data are important in many aspects of disease research, including primary research, target discovery, biomarker identification, and prognostic test determination. The methods used to analyze the data can have a profound effect on the interpretation of the results (17, 18). Network analysis of high-throughput data can be useful in breaking the gap between data production and drug targeting and helps to uncover biological complexity (19, 20). Therefore, to explore the molecular mechanism and discover specific biomarkers for chronic ITP compared with newly diagnosed in pediatrics, we applied bioinformatics techniques to analyze gene expression profiles of pediatric chronic ITP versus newly diagnosed and identify DEGs. For this aim, in the beginning, pediatric chronic ITP patients’ gene expression profiles were compared with pediatric newly diagnosed downloaded from GEO dataset. DEGs were identified using limma packages of the R software. The involvement of DEGs in the biological processes (BP), cellular components (CC), molecular functions (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) were assessed with DAVID online tool. DEGs visualized using Cytoscape software. We applied the network analysis using Cytoscape to predict probable biomarkers. The panther database was used for transcriptional regulatory network construction. These studies could help find crucial genes that might be applied for appropriate diagnostics and treatment strategies in ITP.
Experimental
Microarrays data
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) is a public dataset for storage microarray, and next-generation sequencing data is freely available for users. In this study, ITP genomic data were obtained from GEO with the series accession number GSE46922 and the platform GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array). This dataset included data from thirteen blood samples: seven newly diagnosed and six chronic samples described by Margareta Jernas et al. (21).
Data processing
Data retrieved from GEO were analyzed with R software to discover significantly expressed genes by employing various statistical tests, mainly, the t-statistics and P-value. R software is a fascinating tool to discriminate two or more groups of samples to classify genes, differentially regulated following the same experimental condition. This software can estimate the P-value for significant outcomes by utilizing Limma R packages from the Bioconductor project. Benjamini false discovery rate was concerned in this outline. Here the genes were chosen for more evaluation with P-value < 0.05, and-0.5 >M > 0.5 (M is log2 fold change).
Functional and pathway enrichment analysis
Up-regulated and down-regulated genes were analyzed separately by the DAVID enrichment database (version 6.8) (https://david.ncifcrf.gov). The Database for Annotation, Visualization, and Integrated Discovery (DAVID) is a web-accessible program that provides a comprehensive set of functional annotation tools to disclose the biological meaning behind gene sets. DAVID contains numerous public sources of protein and gene annotation from more than 65,000 species (22). Gene Ontology and KEGG pathway analysis were performed using the DAVID database for functional analysis of the gene lists. We used the functional annotation clustering; to reveal the clusters enriched in gene ontology and KEGG pathway terms with the enrichment score number. Gene Ontology (GO; www.geneontology.org) and the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.ad.jp/KEGG) enrichment analysis were performed to identify DEGs. GO was used for categorization, including biological process, molecular function, and cellular component, which is widely used in bioinformatics and increases the possibility of indentifying the most correlative mechanisms. KEGG was used for understanding the most relevant pathway of informative genes.
Network construction and modules selection
DEGs interaction network can clarify the molecular mechanism of cellular processing. Functional interaction between DEGs was constructed with Cytoscape (version 3.5.1). In this study, the network was extended with the Cytoscape public database. Highly connected nodes were selected as hubs. Some nodes with the highest betweenness centrality were nominated as bottleneck nodes. Then, Molecular Complex Detection (MCODE) was used for screening modules. The functional enrichment analysis of DEGs in each module was performed by DAVID.
Transcriptional regulatory network construction
In order to identify the transcription factor (TF) nodes in the network, the PANTHER Classification System database (http://www.pantherdb.org/) was used (23). Then the transcriptional regulatory network was visualized by Cytoscape.
Results
Data screening
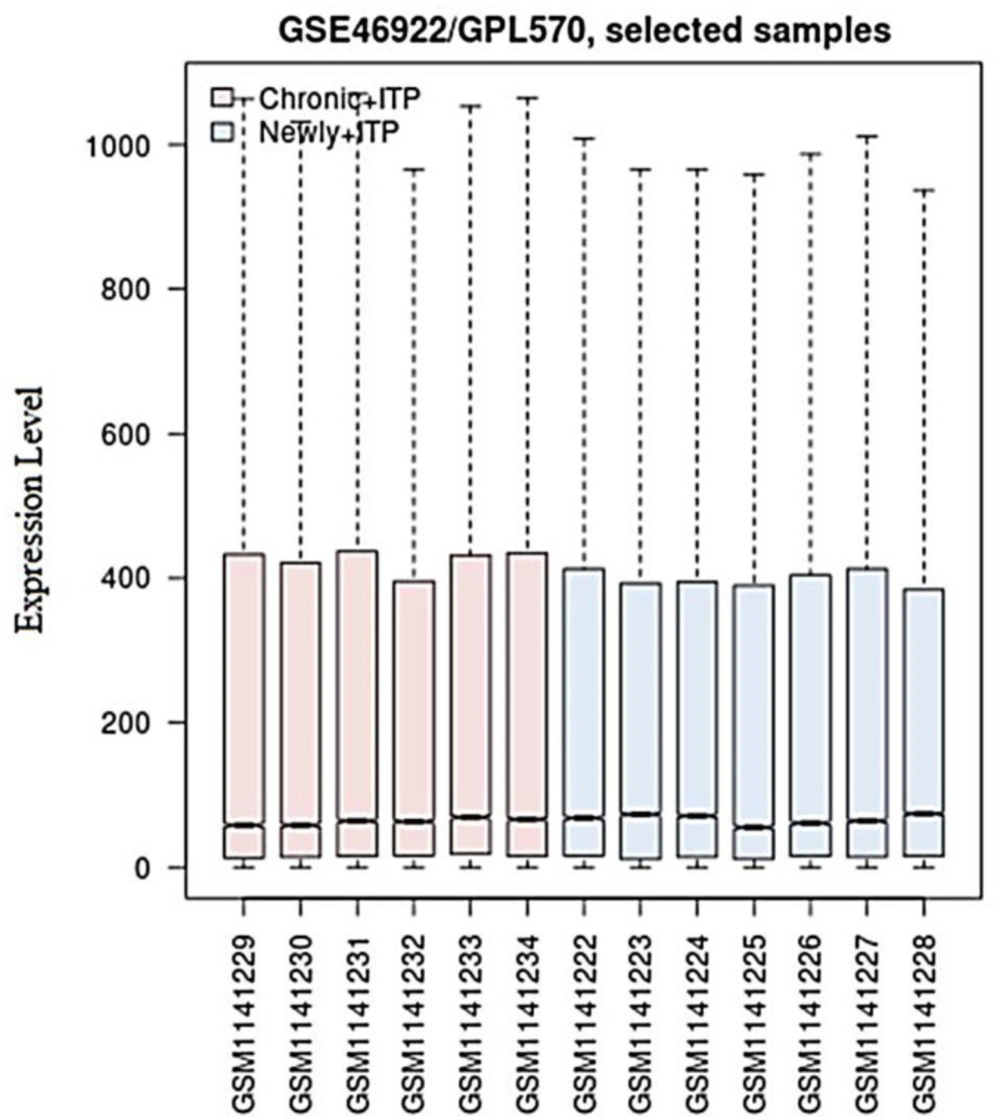
Based on P-value < 0.05 in comparison to newly diagnosed ITP/chronic ITP, 132 DEGs were identified, consisting of 78 up-regulated (Supplementary Table S1) and 54 down-regulated genes (Supplementary Table S2). As shown in (Figure 1), the medians located at the same level after performing data normalization with R software, indicating a perfect effect.
Gene ontology and pathway enrichment analysis
Gene Ontology and Pathway functional enrichment analysis were performed according to the P-values < 0.05 on the identified DEG. The enriched term of BP for up-regulated genes was reported in (Table 1 and Supplementary Table S3). They were significantly involved in 10 significant clusters of biological processes, associated with regulation of autophagy, cell cycle checkpoint, regulation of gene expression, cellular component organization or biogenesis, positive regulation of cell projection organization, macromolecule metabolic process, sister chromatid segregation, the establishment of protein localization to the membrane, and regulation of protein tyrosine kinase activity. The up-regulated genes were located in 5 clusters associated with the intracellular part, organelle, nucleus, intracellular non-membrane-bounded organelle, chromosome, centromeric region, and ciliary membrane (Supplementary Table S4). Significant Molecular function represented in (Supplementary Table S5) involved 2 cluster link to nucleic acid binding and protein kinase activity.
Moreover, the gene ontology related to biological process terms were over-represented in down-regulated DEGs with significant P-value which mainly involved in the modification of morphology or physiology of other organism involved in symbiotic interaction, cellular response to monosaccharide stimulus, programmed cell death, histone methylation, negative regulation of sequence-specific DNA binding transcription factor activity, and positive regulation of binding (Table 2 and Supplementary Table S6).
The significant cellular components, related to down-regulated genes contained four clusters that were mainly involved in intracellular, organelle part, membrane-bounded organelle, lysosomal membrane, lytic vacuole membrane, and chromosomal part. (Supplementary Table S7). Two significant molecular function clusters for down-regulated genes, represented in (Supplementary Table S8), were related to lipase activity, hydrolase activity, acting on ester bonds, and structure-specific DNA binding.
The significant pathway represented in (Table 3) for up-regulated and down-regulated DEGs .the pathway enrichment analysis for up-regulated DEGs indicated these genes involved in the ErbB signaling pathway, mRNA surveillance pathway, and the Estrogen signaling pathway. Whereas, only the Notch signaling pathway is related to down-regulated genes.
Network construction and modules selection
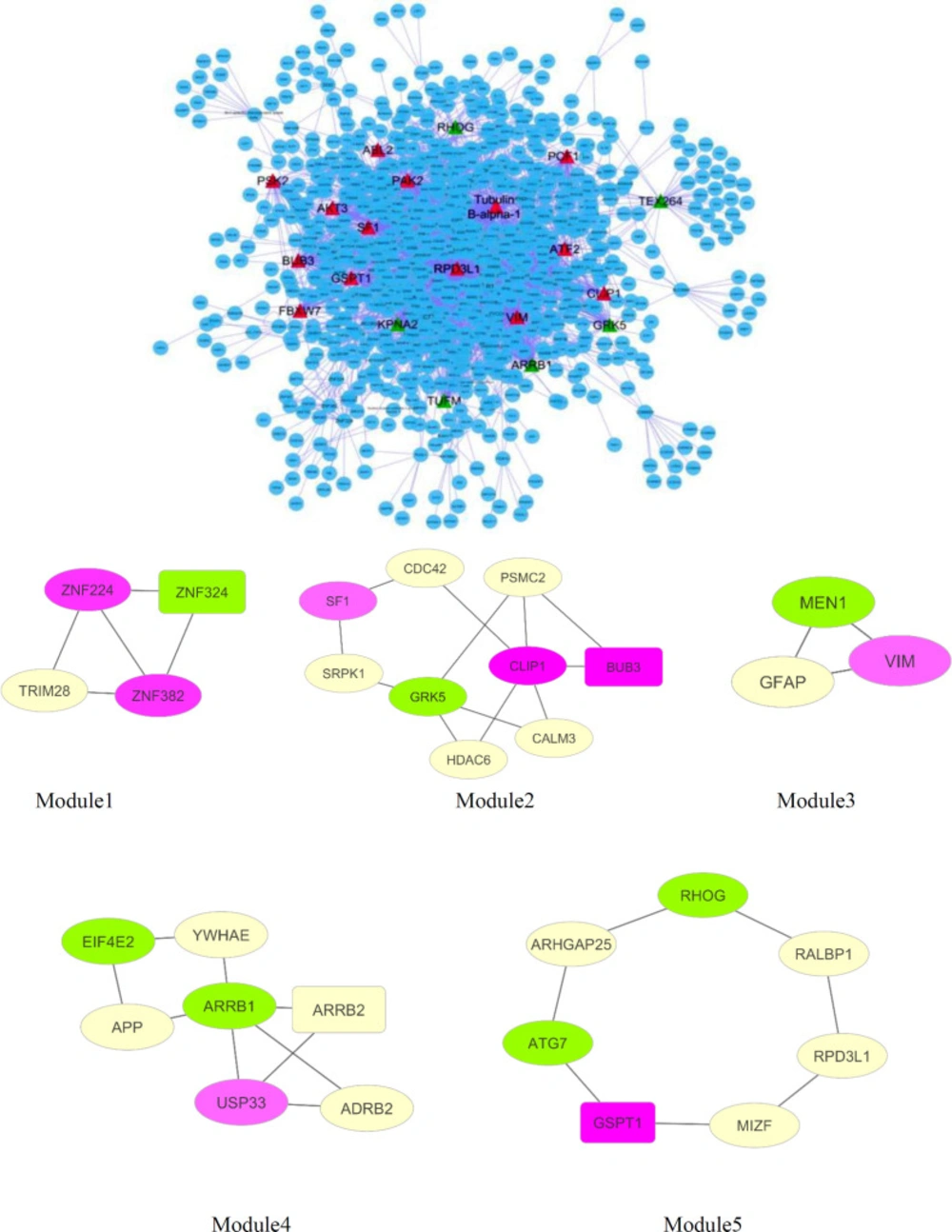
Based on public databases existing on Cytoscape, the PPI network of DEGs was established. Network analysis was shown consisting of 1137 nodes and 2647 edges (Figure.2A). The cut-off criterion of hub gene selection was set at ≥ 40 degrees. Based on this cut-off, twenty hubs are recognized in the network. Therefore, regarding the cut-off criteria, seventeen genes of DEGs were selected as hub nodes. They consisted of eleven up-regulated (ATF2, VIM, PAK2, SF1, BUB3, PCF11, PCF12, FBXW7, GSPT1, CLIP1, ABL2) and six down-regulated (ARRB1, KPNA2, GRK5, TUFM, RHOG, TEX264) genes (Table 4).
Twenty of the highest betweenness centrality including fifteen genes of DEGs were selected as bottleneck containing nine up-regulated (BBS2, RPRD1A, FNBP4, TUBE1, ABCA5, EIF4E3, TNRC6A, TBC1D5, VIM) and six down-regulated (TMEM214, COPRS, MAU2, MRPL45, TBC1D9B, ARRB1) genes (Table 5). Two of the mentioned genes,including VIM and ARRB1, appeared among hub nodes. These two genes were dentified as hub-bottleneck genes. Which confirms the important role of these two genes.
The functional modules were assessed using the MCODE plugin. Five modules were identified including thirty nodes and 36 edges that comprised Module 1 (ZNF324, ZNF224, ZNF382, TRIM28), module 2 (BUB3, CDC42, GRK5, PSMC2, SF1, HDAC6, SRPK1, CLIP1), module 3 (VIM, MEN1, GFAP), module 4 (APP, EIF4E2, ARRB1, ARRB2, ADRB2, YWHAE, USP33), module 5 (ATG7, GSPT1, RALBP1, MIZF, RPD3L1, ARHGAP25, RHOG) (Figure 2B).
We found six DEGs existing in both hub genes and modules, which have significant P-value (P-value < 0.05) for enriched BP. These genes included three up-regulated genes and three down-regulated genes (Table 6).
Transcriptional regulatory network construction:
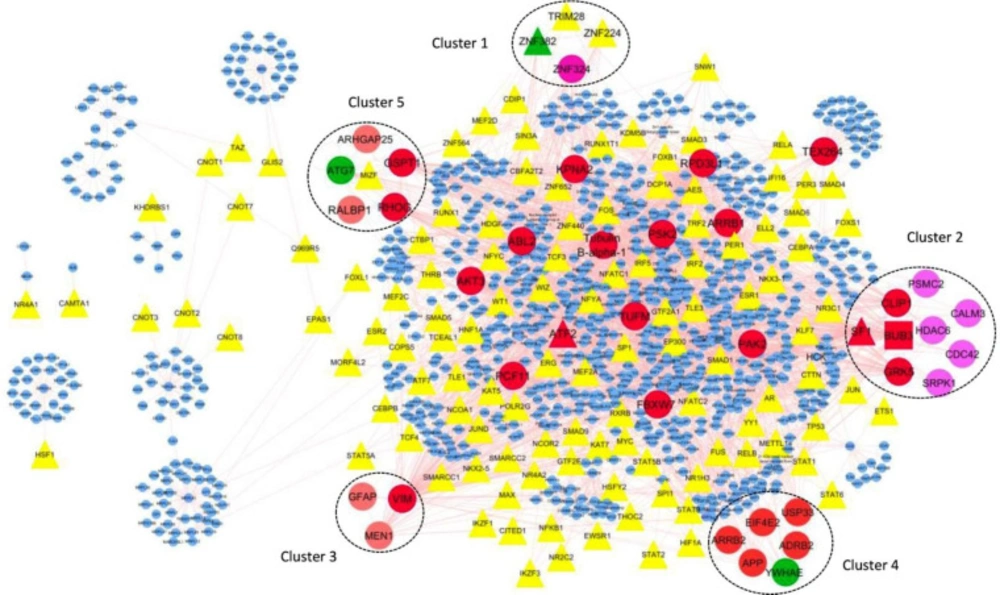
One hundred twenty-one nodes with TF function have been identified from 1137 network nodes using the panther database. By using Cytoscape, these 121 nodes have been visualized in a regulatory network.
Five genes of thirty genes existing in modules are TF, including ZNF224, ZNF382, TRIM28, MIZF, and SF1. ZNF382, TRIM28, ZNF224 belong to module one, SF1 belongs to module two, and MIZF belongs to module 5.
Further analysis of potentially remarkable modules was performed by detecting TFs with a high degree of connections with other nodes, the so-called hub-TFs.
It should be noted that SF1 is the hub node in the transcriptional regulatory network. SF1, as a TF encoding gene is a hub-TF (Figure.3).
Discussion
Diminished platelet production and enhanced platelet destruction are the familiar characters of ITP (24). However, the first hit for dysregulation of the immune system in ITP remains unknown (25). Understanding the molecular and physiopathological mechanisms of ITP requires many efforts to design new preventive and therapeutic strategies. Due to the interaction of genes and environmental factors in common human diseases, a more integrated biological approach is needed to solve these complexities (26). DNA microarrays are used as a powerful technique in biomedical research. This method has attracted much attention from scientists because of its ability to identify thousands of genes and even the entire genome simultaneously (26). Systemic network analysis of high-throughput data is the most useful technique to explain the important implications of life science. Network features, such as composition and topology are highly relevant to vital cellular functions, so they are critical in biological science research (27). This study tries to find essential genes and mechanisms by bioinformatics analysis of GSE46922 microarray data, which are different between the newly diagnosed and the chronic ITP. This study identifies, 131 DEGs, consisting of 78 up-regulated genes and 53 down-regulated genes, which are differentially expressed between, the newly diagnosed ITP and chronic ITP-.
Our enrichment analysis of the up-regulated DEGs showed that autophagy played a significant role in ITP. There is evidence that the positive regulation of autophagy is the most relevant biological process in ITP associated with the expressed genes in the chronic phase. Autophagy induces to the maintenance of platelet life and physiological functions (28). Improper expression of molecules in the autophagy pathway has been also determined in ITP patients lymphocytes (29). Elevating platelet autophagy has been also shown to diminish platelet destruction by prohibiting apoptosis and amending platelet viability (28). Besides, particular evidence implied that megakaryocytes undergo autophagy in ITP patients (30). The apoptotic process was diminished in accordance with activate autophagy process in chronic ITP.
Our study has shown that down-regulated genes in the chronic phase were mainly enriched in the Notch signaling, closely related to hematopoiesis, which involves the evolving hematopoietic system to generate hematopoietic stem cells and the development of immune cells like in T-cells or progress several autoimmune diseases like ITP (32). Rania Mohsen Gawdat et al. found the correlation of Notch1/Hes1 gene expression levels in Egyptian paediatric patients with newly diagnosed and persistent primary ITP (31, 32). We detected this pathway in newly diagnosed ITP while down-regulated in the chronic phase, and this data has shown that the Notch pathway is replaced by the ErbB signaling pathway, mRNA surveillance pathway, and Estrogen signaling pathway over time to display the chronic phase symptom. Also molecular crosstalk among Notch signaling pthway with ErbB and Estrogen signaling pathways was acknowledged in breast cancer (33). This study also confirms the crosstalk between emerging ErbB and Estrogen pathway and inhibition of the Notch signaling pathway in ITP. The mRNA surveillance pathway was enriched by the up-regulated genes related to the quality control mechanism that targets aberrant mRNAs for degradation (34). This pathway was not reported for ITP but confirm this mechanism in autoimmune disease and cellular defense against virus invasion. Mutations affecting the mRNA surveillance machinery cause chronic activation of defense programs, resulting in autoimmune phenotypes. The Systemic lupus erythematosus (SLE) as a human autoinflammatory and autoimmune disorders are notably linked to this system deviation (34). ITP manifests several symptoms of mimicking diseases like SLE; therefore, one might be aware of this similarity emphasizing with several investigations. Besides, this pathway enriched from down-regulated genes in the chronic phase; it implies that the chronic phase of ITP can be due to perturbations in the pathways.
The network analysis also demonstrated that there are interactions among the DEGs.
Our network analysis revealed a set of candidate genes (three up-regulated and three down-regulated) for the investigation of biomarkers or molecular mechanisms of ITP, which was significantly correlated with chronic ITP, including BUB3, GRK5, SF1, VIM, ARRB1, and RHOG.
Our network analysis also verifies the Notch signaling pathway in ITP. In this study, ARRB1 was considered a hub-bottleneck protein with a high degree and high betweenness centrality value. This protein is strongly related to the Notch signaling pathway. Due to its unique features, it has an attractive advantage for drug targeting.
One of the essential genes that play an indispensable role in the maturation of hematopoietic precursors is Vimentin (VIM) that belongs to hub-bottleneck protein. Alteration in expression of VIM has been recognized in the maturation process of the megakaryocytic, granulomonocytic, erythroid, and lymphoid lineages (35). Up-regulated VIM has been also shown in the formation of fully active macrophage-like cells and macrophage polykaryons (36). Rho GTPases (RhoG) is one of the crucial members of our analysis, which has a central regulatory role in platelet production and megakaryocyte maturation (37).
One of the most important genes in this research was SF1. In addition to being a hub, integrating TF’s expression data into Cytoscape indicated that SF1 is also a TF. Kenichi Yoshida et al. reported that there is a mutation in SF1 in hematologic malignancies, but its frequency was not at confidence level for presentation to clinical associations (38).
The G-protein-coupled receptor kinase 5 (GRK5) is a critical member of the threonine/serine kinase family that phosphorylates and regulates the G-protein-coupled receptor (GPCR) signaling pathway. GRK5 has a key role in several diseases; for example, GRK5 is a decisive pathogenic factor in early Alzheimer’s disease, hepatic steatosis and metabolic disorders such as type II diabetes and obesity, injured and failing heart and cancer (39-44). GRK5 also has multiple roles in TLR (Toll-Like Receptor) signaling, which were described as a family of receptors involved in recognizing pathogen-associated molecular patterns (PAMPs) derived from microbes. Moreover, the importance of TLRs has been identified in several inflammatory diseases, including non-infectious diseases (45, 46). In addition, detection of GRK5 expression provides a target for determining the effectiveness of drugs and determining patient prognosis in cancer (47).
The BUB3 is one of the mitotic checkpoint proteins specified by a group of evolutionarily conserved genes. It is believed that the failure of the BUB gene family as a surveillance system is a critical components of the regulatory process which causes genomic instability. This gene family encodes proteins that are a part of a large multi-protein kinetochore complex (48, 49). The BUB3’s importance was found in colorectal cancer at a young age and in low-grade breast cancers (50, 51).
The use of omics technology to identify the mechanism of disease and the discovery of biomarkers has received much attention in recent years. Microarray and proteomics approaches can help to solve biological complexities by creating an extensive list of expressed transcripts that are simultaneously (52). As mentioned in the introduction, Zheng and his colleagues were able to introduce six important markers for the diagnosis of ITP by using Proteomics technology in 2016 (11). However, they have not yet been used in the clinic. Our study using microarray data analysis introduces six new markers that can clarify the pathogenesis of the ITP and need many examinations for clinic application.
(A) network with 1137 nodes and 2647 edges. Unregulated hub genes were shown with red triangle nodes while down-regulated represented with green color (B) Significant modules selected from the network. Pink modules illustrated up-regulated genes, while green nodes illustrated down-regulated genes. Seed nodes are shown in rectangular shape
Visualization in Cytoscape of interactions between TFs, modules and hub-TFs. TFs are shown as triangle. Hubs are displayed in red ellipses. Modules showed by number with different color that contains the nodes are hub (red node), seed (green node) and TF (yellow and red triangle). There is just one red rectangle in module No.2 related to the node that is hub-seed gene. Red triangle related to the nodes are hub-TFs and green triangle is a node related seed-TFs. SF1 and ATF2 are hub-TFs and ZNF382 is a seed-TF. ZNF382 and SF1 are the members of modules No.1 and No.2 respectively
| Category | Term | P-Value | |
|---|---|---|---|
| Annotation Cluster 1 | Enrichment Score: 1.6089173346492356 | ||
| GO:1902589 | single-organism organelle organization | 0.005313 | |
| GO:0000226 | microtubule cytoskeleton organization | 0.020728 | |
| GO:0007017 | microtubule-based process | 0.026109 | |
| Annotation Cluster 2 | Enrichment Score: 1.5454786370206883 | ||
| GO:0010506 | regulation of autophagy | 0.003125 | |
| GO:0010508 | positive regulation of autophagy | 0.03688 | |
| Annotation Cluster 3 | Enrichment Score: 1.1276637957883262 | ||
| GO:0000075 | cell cycle checkpoint | 0.010137 | |
| GO:0022402 | cell cycle process | 0.010174 | |
| GO:0000077 | DNA damage checkpoint | 0.017372 | |
| GO:0007049 | cell cycle | 0.017944 | |
| GO:0031570 | DNA integrity checkpoint | 0.020624 | |
| GO:0007093 | mitotic cell cycle checkpoint | 0.020967 | |
| GO:0045930 | negative regulation of mitotic cell cycle | 0.047204 | |
| Annotation Cluster 4 | Enrichment Score: 1.0524732568167308 | ||
| GO:0016043 | cellular component organization | 0.03475 | |
| GO:0006996 | organelle organization | 0.046543 | |
| GO:0071840 | cellular component organization or biogenesis | 0.04844 | |
| Annotation Cluster 5 | Enrichment Score: 0.9254147029064898 | ||
| GO:0031344 | regulation of cell projection organization | 0.004985 | |
| GO:0010975 | regulation of neuron projection development | 0.018065 | |
| GO:0030030 | cell projection organization | 0.020064 | |
| GO:0031346 | positive regulation of cell projection organization | 0.02586 | |
| GO:0031175 | neuron projection development | 0.030331 | |
| GO:0030182 | neuron differentiation | 0.035135 | |
| Annotation Cluster 6 | Enrichment Score: 0.9097209940740182 | ||
| GO:0043170 | macromolecule metabolic process | 0.001037 | |
| GO:0010468 | regulation of gene expression | 0.008941 | |
| GO:0060255 | regulation of macromolecule metabolic process | 0.008953 | |
| GO:0019222 | regulation of metabolic process | 0.011749 | |
| GO:0044260 | cellular macromolecule metabolic process | 0.014544 | |
| GO:0010467 | gene expression | 0.017106 | |
| GO:0010558 | negative regulation of macromolecule biosynthetic process | 0.033922 | |
| GO:0009892 | negative regulation of metabolic process | 0.038268 | |
| GO:0010605 | negative regulation of macromolecule metabolic process | 0.041577 | |
| GO:0031327 | negative regulation of cellular biosynthetic process | 0.043543 | |
| GO:0031324 | negative regulation of cellular metabolic process | 0.044105 | |
| GO:0009890 | negative regulation of biosynthetic process | 0.047731 | |
| GO:0051172 | negative regulation of nitrogen compound metabolic process | 0.048092 | |
| Annotation Cluster 8 | Enrichment Score: 0.7138986023041735 | ||
| GO:0007062 | sister chromatid cohesion | 0.011798 | |
| GO:0000819 | sister chromatid segregation | 0.049886 | |
| Annotation Cluster 11 | Enrichment Score: 0.5802981069084664 | ||
| GO:0090150 | establishment of protein localization to membrane | 0.046903 | |
| Annotation Cluster 17 | Enrichment Score: 0.42916203800540753 | ||
| GO:0018108 | peptidyl-tyrosine phosphorylation | 0.009263 | |
| GO:0018212 | peptidyl-tyrosine modification | 0.00948 | |
| Annotation Cluster 18 | Enrichment Score: 0.40498640863411367 | ||
| GO:0018108 | peptidyl-tyrosine phosphorylation | 0.009263 | |
| GO:0018212 | peptidyl-tyrosine modification | 0.00948 | |
| GO:0044260 | cellular macromolecule metabolic process | 0.014544 | |
| GO:0061097 | regulation of protein tyrosine kinase activity | 0.023351 | |
| Category | Term | P-Value |
|---|---|---|
| Annotation Cluster 2 | Enrichment Score: 0.7613112520752194 | |
| GO:0044267 | cellular protein metabolic process | 0.011896 |
| GO:0043412 | macromolecule modification | 0.01846 |
| GO:0019538 | protein metabolic process | 0.018791 |
| GO:0006807 | nitrogen compound metabolic process | 0.019514 |
| GO:0009059 | macromolecule biosynthetic process | 0.021932 |
| GO:0043170 | macromolecule metabolic process | 0.028871 |
| GO:0044249 | cellular biosynthetic process | 0.033903 |
| GO:0034641 | cellular nitrogen compound metabolic process | 0.034934 |
| GO:0044237 | cellular metabolic process | 0.043685 |
| GO:0008152 | metabolic process | 0.044952 |
| GO:0044238 | primary metabolic process | 0.04675 |
| GO:0006464 | cellular protein modification process | 0.047376 |
| GO:0036211 | protein modification process | 0.047376 |
| GO:0009058 | biosynthetic process | 0.048575 |
| GO:0071704 | organic substance metabolic process | 0.04992 |
| Annotation Cluster 3 | Enrichment Score: 0.7085147802867421 | |
| GO:0051817 | modification of morphology or physiology of other organism involved in symbiotic interaction | 0.023652 |
| GO:0035821 | modification of morphology or physiology of other organism | 0.033336 |
| Annotation Cluster 5 | Enrichment Score: 0.5438770693729118 | |
| GO:0016570 | histone modification | 0.005122 |
| GO:0016569 | covalent chromatin modification | 0.012772 |
| GO:0006325 | chromatin organization | 0.012912 |
| GO:0090630 | activation of GTPase activity | 0.017421 |
| GO:0032092 | positive regulation of protein binding | 0.017839 |
| GO:0006996 | organelle organization | 0.019368 |
| GO:0018205 | peptidyl-lysine modification | 0.01994 |
| GO:0051817 | modification of morphology or physiology of other organism involved in symbiotic interaction | 0.023652 |
| GO:0071333 | cellular response to glucose stimulus | 0.026068 |
| GO:0071331 | cellular response to hexose stimulus | 0.027564 |
| GO:0071326 | cellular response to monosaccharide stimulus | 0.027564 |
| GO:0051276 | chromosome organization | 0.029444 |
| GO:0006915 | apoptotic process | 0.030391 |
| GO:0071322 | cellular response to carbohydrate stimulus | 0.032253 |
| GO:0035821 | modification of morphology or physiology of other organism | 0.033336 |
| GO:0001678 | cellular glucose homeostasis | 0.034432 |
| GO:0012501 | programmed cell death | 0.042073 |
| GO:0016571 | histone methylation | 0.0443 |
| GO:0006464 | cellular protein modification process | 0.047376 |
| GO:0036211 | protein modification process | 0.047376 |
| GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity | 0.048006 |
| GO:0051099 | positive regulation of binding | 0.0499 |
| Term | Description | P-value | Genes | |
|---|---|---|---|---|
| Up-regulated DEGs | ||||
| 1 | hsa04012 | ErbB signaling pathway | 0.033682 | PAK2, ABL2, AKT3 |
| 2 | hsa03015 | mRNA surveillance pathway | 0.036574 | PCF11, GSPT1, MSI2 |
| 3 | hsa04915 | Estrogen signaling pathway | 0.042636 | FKBP5, AKT3, ATF2 |
| Down-regulated DEGs | ||||
| 1 | hsa04330 | Notch signaling pathway | 0.009201 | HDAC1;MFNG;DTX1 |
| UniProtKB ID | Gene name | Degree | Betweeness centrality | |
|---|---|---|---|---|
| Up-regulated | ||||
| 1 | P15336 | ATF2 | 124 | 0.095148 |
| 2 | P08670 | VIM | 111 | 0.123853* |
| 3 | Q13177 | PAK2 | 101 | 0.07973 |
| 4 | Q15637 | SF1 | 90 | 0.084552 |
| 5 | O43684 | BUB3 | 85 | 0.058294 |
| 6 | O94913 | PCF11 | 77 | 0.037986 |
| 7 | Q9Y243 | PCF12 | 62 | 0.03668 |
| 8 | Q969H0 | FBXW7 | 53 | 0.025345 |
| 9 | P15170 | GSPT1 | 49 | 0.037137 |
| 10 | P30622 | CLIP1 | 46 | 0.012378 |
| 11 | P42684 | ABL2 | 42 | 0.023944 |
| Down-regulated | ||||
| 1 | P49407 | ARRB1 | 125 | 0.121072* |
| 2 | P52292 | KPNA2 | 81 | 0.066201 |
| 3 | P34947 | GRK5 | 59 | 0.036356 |
| 4 | P49411 | TUFM | 56 | 0.035678 |
| 5 | P84095 | RHOG | 40 | 0.028479 |
| 6 | Q9Y6I9 | TEX264 | 40 | 0.055543 |
| Added by network | ||||
| 1 | Q13547 | RPD3L1 | 202 | 0.236783* |
| 2 | Q71U36 | Tubulin B-alpha-1 | 134 | 0.135022* |
| 3 | Q7L7X3 | PSK2 | 42 | 0.017018 |
| UniProtKB ID | Gene name | Betweenness centrality | Degree | |
|---|---|---|---|---|
| Up-regulated | ||||
| 1 | Q9BXC9 | BBS2 | 1 | 27 |
| 2 | Q96P16 | RPRD1A | 1 | 24 |
| 3 | Q8N3X1 | FNBP4 | 1 | 6 |
| 4 | Q9UJT0 | TUBE1 | 1 | 2 |
| 5 | Q8WWZ7 | ABCA5 | 1 | 2 |
| 6 | Q8N5X7 | EIF4E3 | 1 | 2 |
| 7 | Q8NDV7 | TNRC6A | 0.44370861 | 27 |
| 8 | Q92609 | TBC1D5 | 0.48979592 | 11 |
| 9 | P08670 | VIM | 0.41453744 | 111* |
| Down-regulated | ||||
| 1 | Q6NUQ4 | TMEM214 | 1 | 13 |
| 2 | Q9NQ92 | COPRS | 1 | 8 |
| 3 | Q9Y6X3 | MAU2 | 1 | 5 |
| 4 | A0A087X2D5 | MRPL45 | 0.807327 | 37 |
| 5 | Q66K14 | TBC1D9B | 0.76268116 | 14 |
| 6 | P49407 | ARRB1 | 0.12107173 | 125* |
| Added by network | ||||
| 1 | Q13618 | CUL3 | 0.48846676 | 2 |
| 2 | Q13547 | RPD3L1 | 0.23678326 | 202* |
| 3 | Q9H492 | MAP1LC3A | 0.23550725 | 2 |
| 4 | Q9H0R8 | GABARAPL1 | 0.23550725 | 2 |
| 5 | Q71U36 | Tubulin B-alpha-1 | 0.13502234 | 134* |
| Gene name | Gene ID | Degree | Betweeness centrality | M | Biological process | |
|---|---|---|---|---|---|---|
| Up-regulated | ||||||
| 1 | VIM | P08670 | 111 | 0.12385346 | 1.077912156 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process (GO:2000060) |
| 2 | SF1 | Q15637 | 90 | 0.08455183 | 2.419055031 | mRNA splice site selection (GO:0006376),spliceosomal complex assembly (GO:0000245),mRNA 3'-splice site recognition (GO:0000389) |
| 3 | BUB3 | O43684 | 85 | 0.05829374 | 0.846438817 | regulation of translation (GO:0006417) |
| Down-regulated | ||||||
| 1 | ARRB1 | P49407 | 125 | 0.12107173 | -1.855926621 | regulation of Notch signaling pathway (GO:0008593), negative regulation of sequence-specific DNA binding transcription factor activity (GO:0043433), negative regulation of NF-kappaB transcription factor activity (GO:0032088), positive regulation of histone H4 acetylation (GO:0090240), desensitization of G-protein coupled receptor protein signaling pathway (GO:0002029), regulation of histone H4 acetylation (GO:0090239), positive regulation of cellular metabolic process (GO:0031325), contractile actin filament bundle assembly (GO:0030038), stress fiber assembly (GO:0043149), negative regulation of cytokine production (GO:0001818), positive regulation of peptidyl-lysine acetylation (GO:2000758), negative regulation of interleukin-8 production (GO:0032717), modification-dependent protein catabolic process (GO:0019941) |
| 2 | GRK5 | P34947 | 59 | 0.03635575 | -0.896145434 | tachykinin receptor signaling pathway (GO:0007217), regulation of signal transduction (GO:0009966), positive regulation of cell proliferation (GO:0008284), regulation of cell proliferation (GO:0042127) |
| 3 | RHOG | P84095 | 40 | 0.02847864 | -1.37279767 | Rac protein signal transduction (GO:0016601), activation of GTPase activity (GO:0090630), positive regulation of GTPase activity (GO:0043547), positive regulation of cell proliferation (GO:0008284), engulfment of apoptotic cell (GO:0043652), phagocytosis, engulfment (GO:0006911), neutrophil degranulation (GO:0043312),neutrophil activation involved in immune response (GO:0002283), neutrophil mediated immunity (GO:0002446), regulation of cell proliferation (GO:0042127) |
Conclusion
The current study has obtained DEGs using comprehensive bioinformatics analysis of high-throughput data released from microarray analysis to find the possible biomarkers. In summary, a total of 132 DEGs were screened, and six genes, including BUB3, GRK5, SF1, VIM, ARRB1, and RHOG, previously have not been reported as signature genes in ITP; here we found that they might play critical roles in chronic ITP. This research contributes new insights into the molecular mechanisms of newly diagnosed ITP and chronic ITP. These six genes together could be considered as a panel of biomarkers to differentiate newly from chronic ITP. Thus, additional investigations are needed to focus on the clinical application of these genes.