1. Background
APIs have been developed from low molecular weight drugs to peptides, proteins, and antibodies in recent years (1). The use of biopharmaceuticals, such as peptide and protein drugs, for the treatment of various diseases is quickly expanding (2). The therapeutic effectiveness of these products is due to their enhanced selectivity and potency, longer duration of action (delayed clearance from the body), and decreased intrinsic toxicity (3, 4). These features provide a distinct advantage over low molecular weight drugs, frequently associated with off-target effects and toxic metabolites (3). However, the efficacy and potency of therapeutic proteins are challenged by several factors, such as stability, blood circulation time, and specificity. Some of these limitations for therapeutic proteins have been overcome by creating bioconjugates through the covalently linking polymers to proteins (5).
Water soluble polyethylene glycols (PEGs) have been extensively employed to chemically modify the surface of protein macromolecules to inhibit their enzymatic breakdown and clearance by the kidneys. PEG binds to some specific surface areas of protein and prevents its hydrolysis without generating significant adverse effects (6). Synthesis of protein-polymer conjugates (so-called PEGylation process) exhibits a unique combination of properties derived from both biological and synthetic materials, which can be tailored to achieve the desired effect (7). Proteins are sensitive, and when chemically modified, their secondary structures should not be altered in a way that affects their activity. However, despite the ability of PEG to improve protein stability, PEGylation has drawbacks, such as significantly lowering the titer and bioactivity of proteins (8).
An alternative approach for protein delivery and enhancing in vivo stability of physiologically active proteins is Supramolecular (self-assembly PEGylation retaining activity; SPRA) technology. Using supramolecular chemistry in the design of biomaterials based on host-guest interactions offers significant advantages due to direct molecular recognition between host-guest pairings and the variety of interaction strengths that can be achieved between different pairs (9). The concept of host-guest supramolecular chemistry is based on creating a noncovalent complex between a guest molecule and the portal of a macrocyclic cavitand (10). To establish a good fit and produce a thermodynamically stable complex, the physicochemical properties of the host cavity (e.g., size, shape, charge, hydrophobicity) and the guest molecule must be complementary (11). Over the last three decades, many host molecules, mainly composed of macrocyclic molecules, have been synthesized (9). Among the synthetic macrocyclic hosts, cyclodextrins (CDs, and in particular β-CD) are commonly utilized due to their excellent biocompatibility, low cytotoxicity, and biodegradability (12).
The ability of CDs to form selective host-guest inclusion complexes depends on the guest molecule appropriately fitting into the CD cavity (13-15).
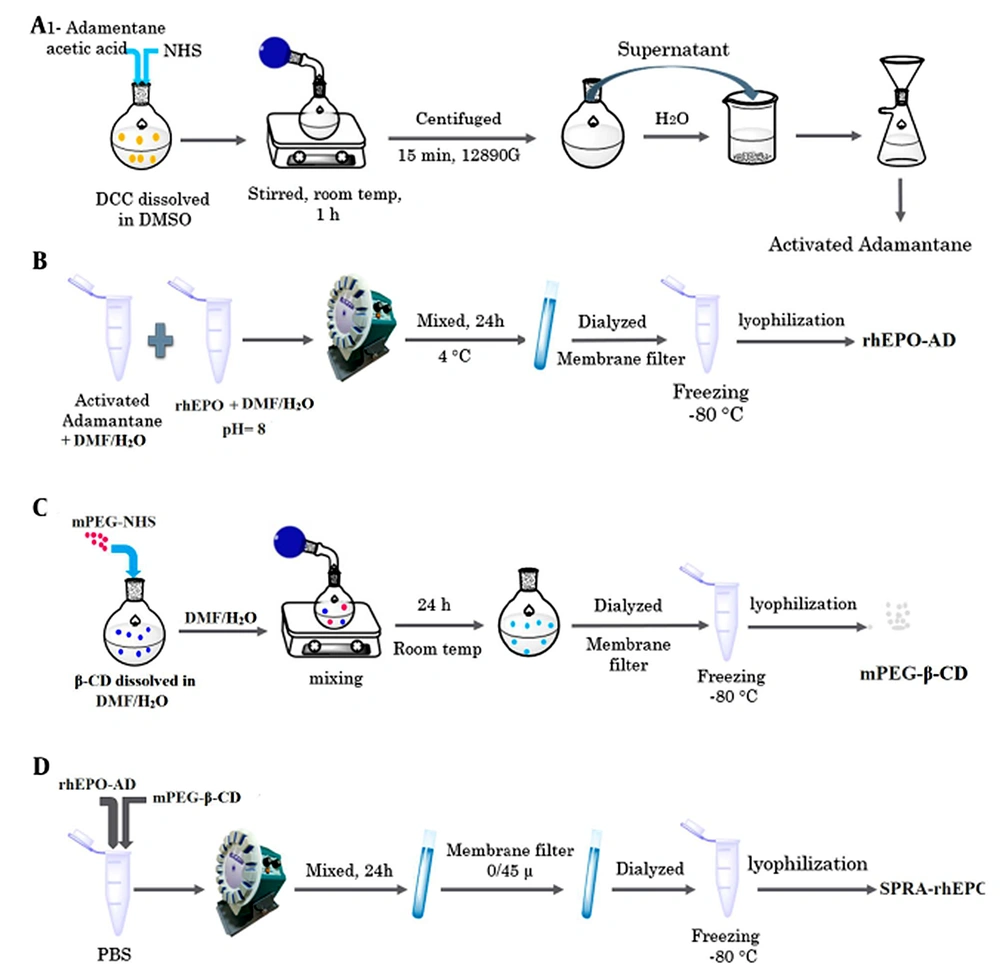
Recombinant human erythropoietin (rhEPO) is one of the most effective proteins for anemia in individuals with chronic renal disease (16). rhEPO has a short half-life, and the PEGylation approach only enhances its biological activity by 4% (17, 18). The main goal of this project was to employ the SPRA technology as a novel approach to lengthen the protein half-life and activity. In the first part of our investigation, we reported the synthesis of the SPRA-rhEPO inclusion complex and employed various analytical techniques to confirm the preparation of the complex (Alizadeh et al. unpublished work). we used adamantane (AD), which plays the role of a guest molecule for β-CD and a linker for rhEPO. Briefly, the SPRA-rhEPO synthesis involves a three-phase reaction, namely the preparation of AD-rhEPO conjugate, the PEGylation of β-CD, and the formation of the PEG-β-CD-AD-rhEPO complex. As known, proteins need to be handled carefully because of their chemical and physical instabilities. Therefore, the secondary structure of rhEPO has to be analyzed to determine the possibility of denaturation during the various synthesis steps.
2. Objectives
This work represents the investigation into the secondary structure of rhEPO, alone, before and after the conjugation reaction with AD, and in the resultant inclusion complex. Various analytical techniques were employed for this purpose, and the protein thermal stability was also evaluated.
3. Methods
3.1. Materials
Recombinant human erythropoietin (rhEPO) solution (in citrate buffer) was supplied by Dorsa Pharmaceutical Co. (Tehran, Iran). 1-adamantane acetic acid (AD-CH2COOH), N-hydroxysuccinimide (NHS), N, N' dicyclohexylcarbodiimide (DCC), dimethylformamide (DMF), dimethyl sulfoxide (DMSO), mono-(6-amino-6-deoxy)-β-cyclodextrin (6-NH2-β-CD) and mPEG 20K-succinimidyl carboxymethyl ester (mPEG-NHS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). All chemicals and solvents were analytical grades. Deionized double-distilled water was utilized throughout this study. No further purification was done on any compound.
3.2. Preparation of SPRA-rhEPO Inclusion Complex
Synthesis of the SPRA-rhEPO complex has been reported in our previous investigation (Alizadeh et al. unpublished work). The complex (mPEG-β-CD-AD-rhEPO) was prepared by forming an inclusion complex between the AD-rhEPO conjugate and the mPEG-β-CD moiety via host-guest interactions. For the synthesis of the guest molecule (i.e., AD-rhEPO), the carboxyl group of AD-CH2COOH (190 mg) was activated by reacting with NHS (230 mg) and DCC (820 mg) in DMSO (6 mL). The solution was stirred for 1 h at 24°C and centrifuged at 12,890 g for 15 min to eliminate the byproducts (e.g., dicyclohexylurea). The activated AD was then precipitated by adding the supernatant to 300 mL of water, and the resultant precipitate was dried under reduced pressure. rhEPO and the activated AD (with a molar ratio of 1:1) were dissolved in 5 ml DMF/distilled water (3:2 v/v, pH 8), and the mixture was stirred at 4°C for 24 h. The reaction solution was lyophilized (Alpha 1-2 LDplus, Germany) after being dialyzed using a membrane filter (Spectra/Por® membrane MWCO: 12 kDa).
For the synthesis of mPEG-β-CD moiety as the host molecule, mPEG-NHS (1 g) was reacted with 6-NH2-β-CD (590 mg) in 20 mL DMF/distilled water (3:2 v/v). The reaction solution was stirred for 24 h at room temperature under vacuum conditions and dialyzed for three days with a membrane filter (Spectra/Por® membrane MWCO: 8 kDa). After removing the unreacted 6-NH2-β-CD, the sample was lyophilized (Alpha 1-2 LD plus, Germany), and the mPEG-β-CD powder was collected.
In the final step, a solution of AD-rhEPO (0.1 mM) and a solution of mPEG-β-CD (0.1 mM) were prepared separately in phosphate buffer saline (PBS, pH 7.4). These solutions were mixed and agitated for 24 h at 24°C to obtain a clear solution. The resultant solution was filtered (0.45 µm syringe filter; MS® PES Syringe Filter) to eliminate the undissolved particles, frozen and freeze-dried. The synthetic pathway is schematically represented in Figure 1.
3.3. SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was carried out in accordance with Laemmli's methodology (19). rhEPO (solution, pH 7.4), rhEPO/DMF solution (pH 8), and AD-rhEPO in citrate buffer were loaded on polyacrylamide gels (4% stacking gel, 12.5% running gel). As standards, 116, 70, 45, 35, 25, 18.4, and 14.4 kDa unstained protein molecular weight markers were utilized. Before loading on the gel, each sample (as a solution in citrate buffer) was heated for 10 - 30 min at 100°C in Laemmli buffer (62.5 mM Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, and 0.01% bromophenol blue). The gel electrophoresis well was loaded with 10 - 12 µL of each sample. The experiment was run under reducing conditions (using β-mercaptoethanol) at a constant current of 20 - 30 mA. After 2 h, the possible aggregated protein and/or its fragments were separated based on their molecular weights. To detect the protein bands, the gel was dyed with a 1% solution of Coomassie brilliant blue R-250.
3.4. Attenuated Total Reflection - Fourier Transform Infrared Spectroscopy
rhEPO and lyophilized rhEPO samples were studied by attenuated total reflection (ATR) - Fourier transform infrared spectroscopy (FTIR). rhEPO injection solution containing citrate buffer (20 mM, pH 7.4) was dialyzed through a membrane filter (Spectra/Por® membrane MWCO: 12 kDa) at 4°C for 48 h. The solution was filtered through a 0.2 µm syringe filter (MS® PES Syringe Filter, Sigma–Aldrich), stored at -80°C, and then lyophilized in a freeze dryer (Alpha 1-2 LDplus, Germany). The conservative lyophilization cycle involved first pre-cooling to -2°C (15 min) as the first step, followed by freezing at -40°C (50 min) and drying under vacuum (70 mTorr) at -35°C for 10 h, -20°C for 8 h, -5°C for 6 h, with continuous drying (100 mTorr) at 10°C for 6 h, 25°C for 6 h and 4°C for 0.5 h (20). There was no attempt to optimize the cycle for rhEPO. As a non-lyophilized control sample, rhEPO solution in citrate buffer was diluted (with citrate buffer) to 1.0 mg/mL. The lyophilized powder was also reconstituted in the citrate buffer to obtain a final rhEPO concentration of 1.0 mg/mL. A sufficient amount of both reconstituted and control solutions were used to analyze with ATR-FTIR (Agilent Technologies Cary 630 FT-IR spectrophotometer with Diamond ATR accessory, Agilent, USA). Dry air was continually purged into the sample chamber, housing the ATR cell. Sixty-four scans were obtained at a resolution of 4 cm-1 in the 4000 - 400 cm-1 range. Bio-RAD® software was used to collect and process the data.
3.5. FTIR
FTIR spectroscopy was carried out using an Agilent Technologies Cary 630 FTIR spectrometer (Agilent, USA) with an average of 64 scans at 4 cm-1 resolution from 4000 to 400 cm-1 at room temperature. To prepare the samples for analysis, rhEPO and AD-rhEPO powders were mixed with KBr (with a ratio of 1:100), and the data was collected and processed using Bio-RAD® software.
3.6. Circular Dichroism Spectroscopy
Circular Dichroism Spectroscopy (CD) spectra of rhEPO and AD-rhEPO powder samples were collected at 25°C with a J-810 Spectrometer (Jasco, Japan) at the wavelength range of 190 - 350 nm. Distilled water was used to prepare solutions with a concentration of 0.2 mg/mL. For the measurement in the far-UV area, the path length of the cuvette was 1 mm. After scanning each spectrum three times, the average spectrum was plotted. Far-UV-CD measurements were performed to investigate the effect of the linker (AD) on the secondary structure of rhEPO.
3.7. Thermal Stability
Thermal stability studies were carried out on SPRA-rhEPO complex, AD-rhEPO, and rhEPO samples (1 mg/mL in phosphate buffer, pH 7.4). The solutions were incubated at 37°C for ten days. At the relevant times, samples were collected and centrifuged (18,000 g for 10 min). A Nanodrop spectrophotometer (Thermo Fisher, USA) was used to measure the absorbance of the supernatant at 280 nm.
4. Results and Discussion
4.1. SDS-PAGE Analysis
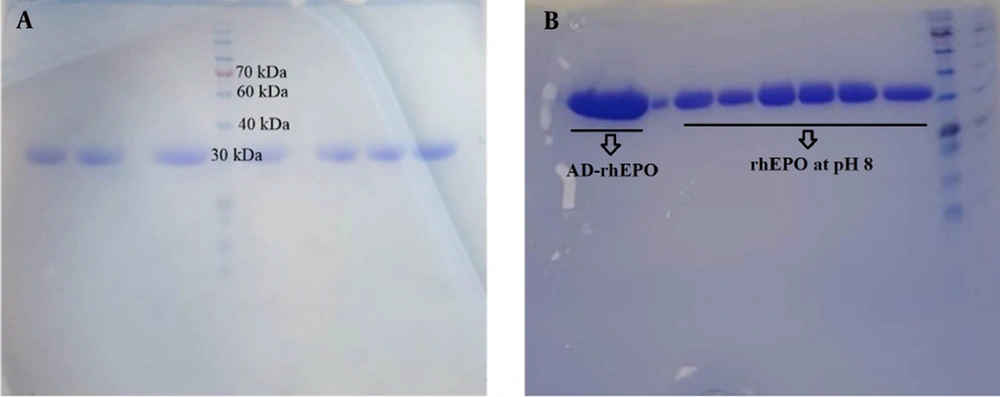
SDS-PAGE is a method commonly used to separate proteins only based on the differences in their molecular weight. This study evaluated the molecular weight and the relevant stability of the protein and its conjugate with AD. rhEPO was treated with DMF, and pH was adjusted to 8 to follow the binding of the AD to rhEPO. Figure 2A illustrates a single band around 30 kDa for rhEPO, and Figure 2B depicts the SDS-PAGE analysis of rhEPO at pH 8 and after binding AD to rhEPO, indicating single bands comparable to the native rhEPO. There was no evidence of any aggregate or structural disruption in the samples, suggesting that neither pH adjustment nor covalent bond would affect the structural integrity of the protein.
4.2. FTIR Analysis
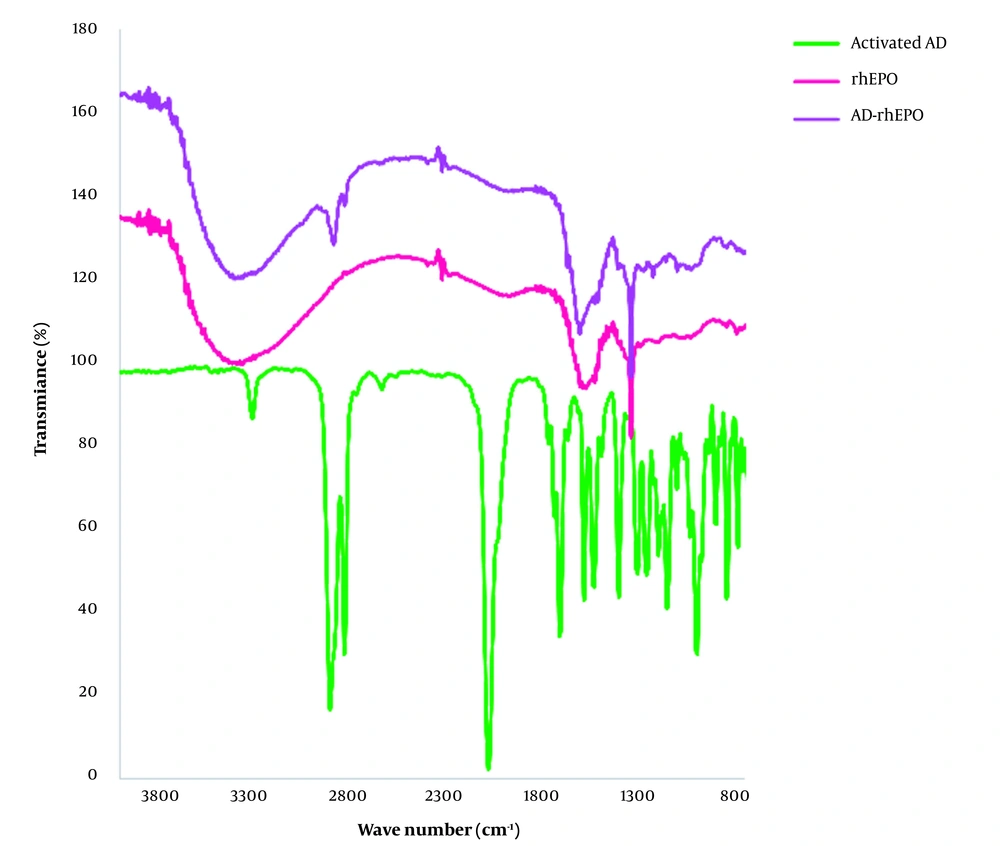
FTIR spectroscopy is a nondestructive technique to determine the structural properties of proteins (21). Multiple vibrational frequencies are present in protein molecules. The polypeptide repeat unit gives rise to 9 distinct group frequencies, namely as amide A (~3,300 cm-1), amide B (~3,100 cm-1), amide I (~1,650 cm-1), amide II (~1,550 cm-1), amide III (~1,300 cm-1), amide IV (~735 cm-1), amide V (~635 cm-1), amide VI (~600 cm-1) and amide VII (~200 cm-1) (22). Vibration frequencies can be associated with individual secondary structural folding due to the differential pattern in H-bonding and geometric orientations of amide bonds in α-helices, β-sheets, β-turn, and random coil structures (23). The two prominent bands in the protein IR spectrum are related to the amide I and amide II bands. The secondary structure of proteins and polypeptides has been extensively evaluated using the amide I band (24). The C=O stretching vibration of the amide group, along with the in-phase bending of the N-H bond and stretching of the C-N bond, are the sources of the amide I. Each frequency of these vibrations, which range between 1,700 and 1,600 cm-1, corresponds to a particular structure and is closely tied to the backbone conformation.
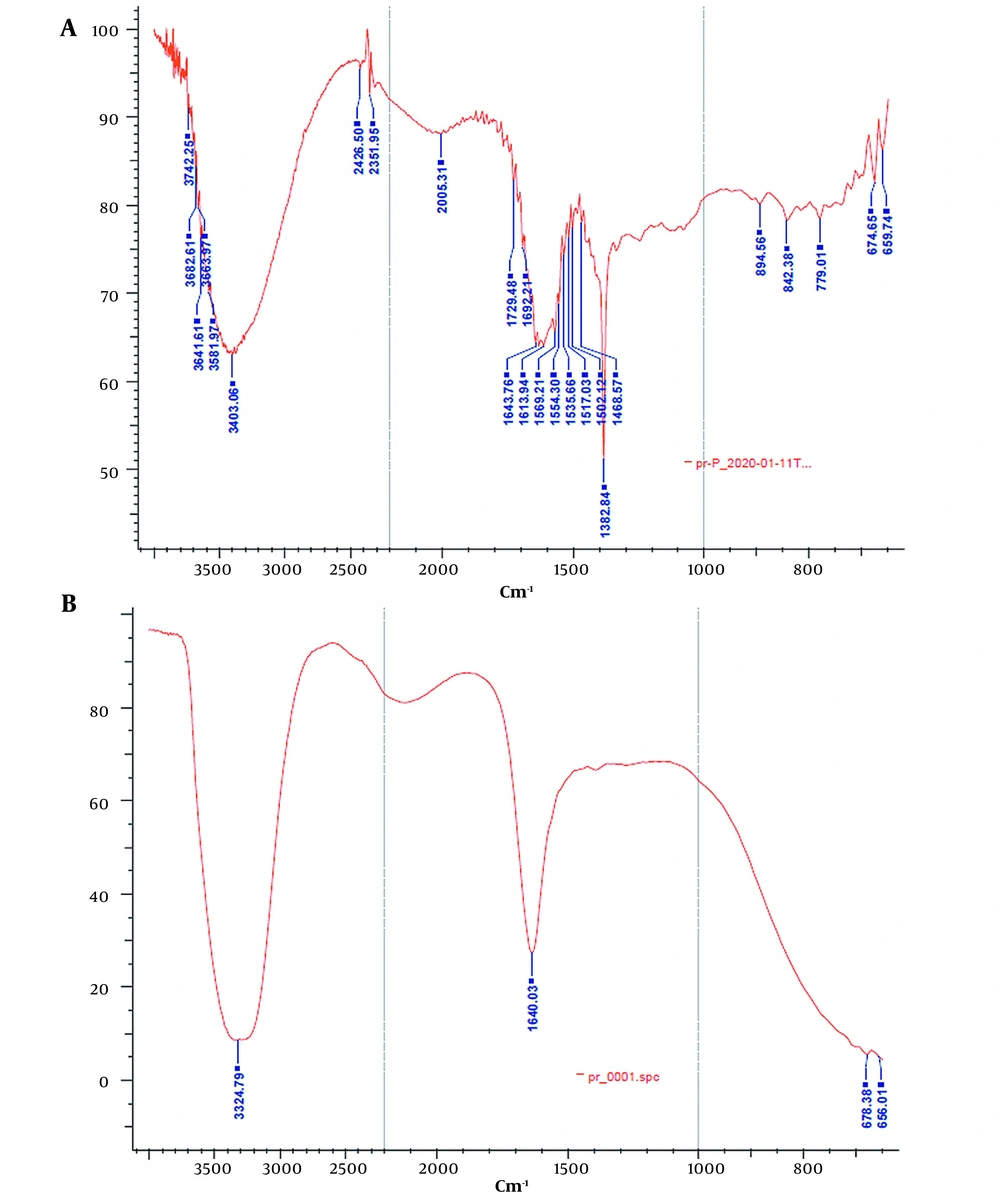
ATR-FTIR analysis was performed to investigate the effect of the lyophilization process on the secondary structure of rhEPO. The ATR-FTIR spectra of the lyophilized rhEPO and rhEPO samples are shown in Figure 3. In Figure 3A, the peaks in the region of 3403.0 cm-1 and 1692.2 cm-1 correspond to the N-H stretch and β-turn structure, respectively. Alpha helix, beta planes, and the side chain structures were characterized by the peaks in the regions of 1643.7 cm-1, 1620.0 cm-1, and 1613.9 cm-1, respectively. In the ATR spectrum of the lyophilized rhEPO (Figure 3B), the peak in the region of 3324.7 cm-1 corresponds to amide A, and the peak in the region of 1640.0 cm-1 corresponds to amide I. The typical amide I bands are located at a wavelength of approximately 1640 cm-1, confirming that the protein possesses an antiparallel ß-sheet secondary structure. Based on the results, it was concluded that rhEPO and the lyophilized rhEPO have the same secondary structures.
FTIR spectra of rhEPO, AD-rhEPO conjugate, and the activated AD are presented in Figure 4. For AD-rhEPO, the band at 2907.3 cm-1 represents the -C-H groups of AD, and the absorption band at 1645 cm-1 corresponds to amide I. By comparison of the FTIR spectra of rhEPO and AD-rhEPO samples, it seemed that the secondary structure of rhEPO was preserved during the reaction with AD, supporting the CD result. The peak corresponding to the CH group of AD was also observed in AD-rhEPO spectra.
4.3. Circular Dichroism Spectra
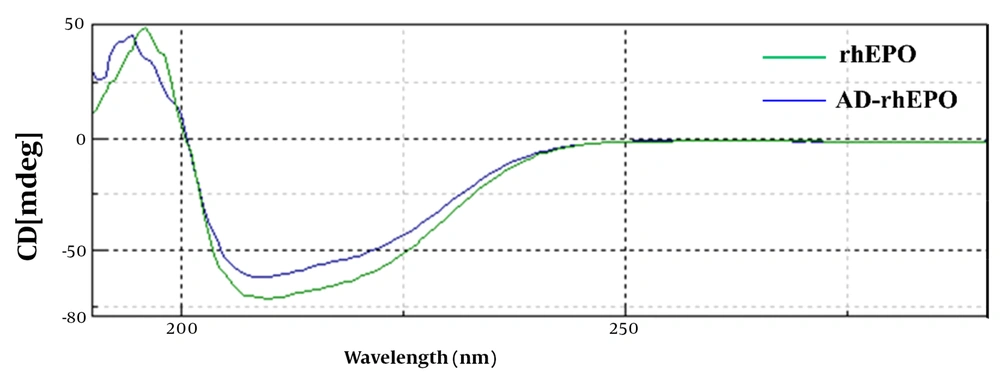
Far-UV circular dichroism was examined to determine the secondary structural changes of rhEPO during reaction with AD. As shown in Figure 5, the CD profile of AD-rhEPO conjugate was comparable to that of rhEPO form with distinctive minima observed at 208 and 217 nm, demonstrating that rhEPO and AD-rhEPO have the same secondary structures, and this confirms the presence of the helix structure. Data of the secondary structure analysis of rhEPO and AD-rhEPO extracted from the CD spectra is depicted in Table 1. According to Greenfield and Fasman, all parameters were calculated (25). In all cases (α-helix, β-sheet, β-turn, Random coil) of the AD-rhEPO, a negligible amount of change was observed compared to that of rhEPO alone, suggesting that rhEPO in the conjugate with AD-rhEPO retains its secondary structure.
| Sample | α-helix | β-sheet | β-turn | Random Coil |
|---|---|---|---|---|
| rhEPO | 19.8% | 38.3% | 13% | 28.9% |
| AD-rhEPO | 18.6% | 39.4% | 14.4% | 27.6% |
4.4. Thermal Stability
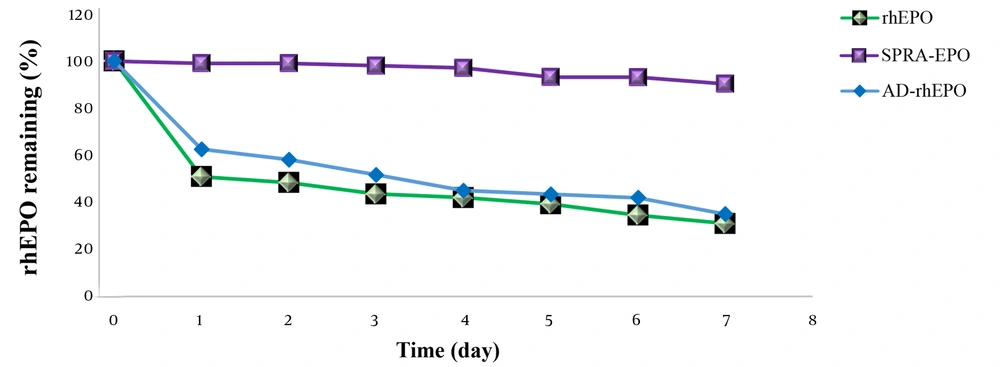
To examine the thermal stability of rhEPO, AD-rhEPO, and SPRA-rhEPO complex, the samples were first dissolved in the same buffer and incubated at 37°C. The residual rhEPO was then measured (Figure 6). Approximately 50% of rhEPO in rhEPO solution or AD-rhEPO was aggregated within one day, whereas the SPRA-rhEPO complex demonstrated high thermal stability compared to the protein alone. These results were in accordance with the results reported by Hirotsu et al. (17).
4.5. Conclusions
We proposed the application of a novel technology, the so-called SPRA technology, for the delivery of rhEPO and synthesized an SPRA-rhEPO inclusion complex. The main goal of this proposal was to improve the half-life and bioactivity of rhEPO. In the first part of this project, we reported the synthesis of the SPRA-rhEPO inclusion complex with β-cyclodextrin and presented the evidence obtained by analytical techniques that confirmed the complexation. In this study, as the complementary part of the previous investigation, we focused on the stability of the protein when it undergoes the synthetic reactions that lead to the formation of the complex. Results obtained from the analytical methods revealed that the stability of the protein was not affected by the reaction conditions. This study highlighted the potential of the SPRA technology as a new strategy for the manipulation of proteins in an attempt to improve protein stability.