1. Background
The most prevalent and severe brain cancer in adults is glioma (1). The kind, location, and severity of the tumor and the patient's general health determine the treatment options for acute glioma (2). Poor treatment and frequent recurrence of glioma result from the blood-brain barrier, which limits the entry of chemical medications into the brain, and the tumor's recurrence is due to the self-renewal of glioma stem cells. The average survival time for glioma patients has decreased due to this phenomenon (3).
Cancer develops when cells accumulate genetic mutations over time. Additionally, some genetic modifications, such as driver mutations, can create cancer cells (4). The majority of the time, treatment plans are based on histology subgroups. However, in addition to the traditional histological categories, each form of cancer can now be further broken down into many molecular subgroups that are crucial in the treatment choice (5). Medical professionals can forecast treatment outcomes and patient survival and treat each genomic subtype differently. As a result, treatment plans are changed in accordance with new molecular markers. Therefore, identifying new treatment options is critical for improving patient survival and clinical outcomes (6).
Telomerase is rapidly activated in germ cells, hematopoietic cells, stem cells, as well as highly reproducible cells. In contrast, telomerase activity in somatic cells is very low, mostly due to the strong regulation of hTERT (7). A deeper knowledge of the underlying mechanisms of hTERT regulation has recently gained momentum, mostly due to advancements in the detection of hTERT promoter mutations. Over the past 20 years, researchers have studied the processes that control hTERT. Nevertheless, additional alterations have been found and shown to increase hTERT expression through a number of genetic and epigenetic mechanisms, such as hTERT amplification, hTERT structural mutations, hTERT promoter mutations, and epithelial alterations genetically regulated by hTERT promoter methylation (8).
Although some researchers have demonstrated hypomethylation in the CpG islets surrounding the hTERT promoter, others have reported increased DNA methylation in cancer cells that express hTERT (9). As a matter of fact, hTERT was among the first genes in which promoter methylation and gene expression had a favorable association. This association between the hTERT promoter's methylation and telomerase activity and hTERT mRNA shows that the hTERT promoter may regulate this gene, but not in the same manner that promoter methylation regulates other genes (10).
Biotechnology and genetic engineering have undergone a significant transformation in recent years thanks to practical and affordable gene editing technologies like CRISPR/Cas9. Genomic alterations can be targeted and employed to cure hereditary illnesses using the CRISPR/Cas9 system (11). Methylation modification is one method of gene editing available with the CRISPR/Cas9 system. The DNMT enzyme family is responsible for DNA methylation (12). Human DNA methyltransferase's catalytic domain performs an enzymatic role in transfected cells, making DNMT3A the most active isoform (13).
2. Objectives
In this study, the main promoter of the hTERT gene and its expression in the U87 cell line for malignant gliomas were targeted using CRISPR/Cas9 system.
3. Methods
3.1. Plasmid Vector Preparation
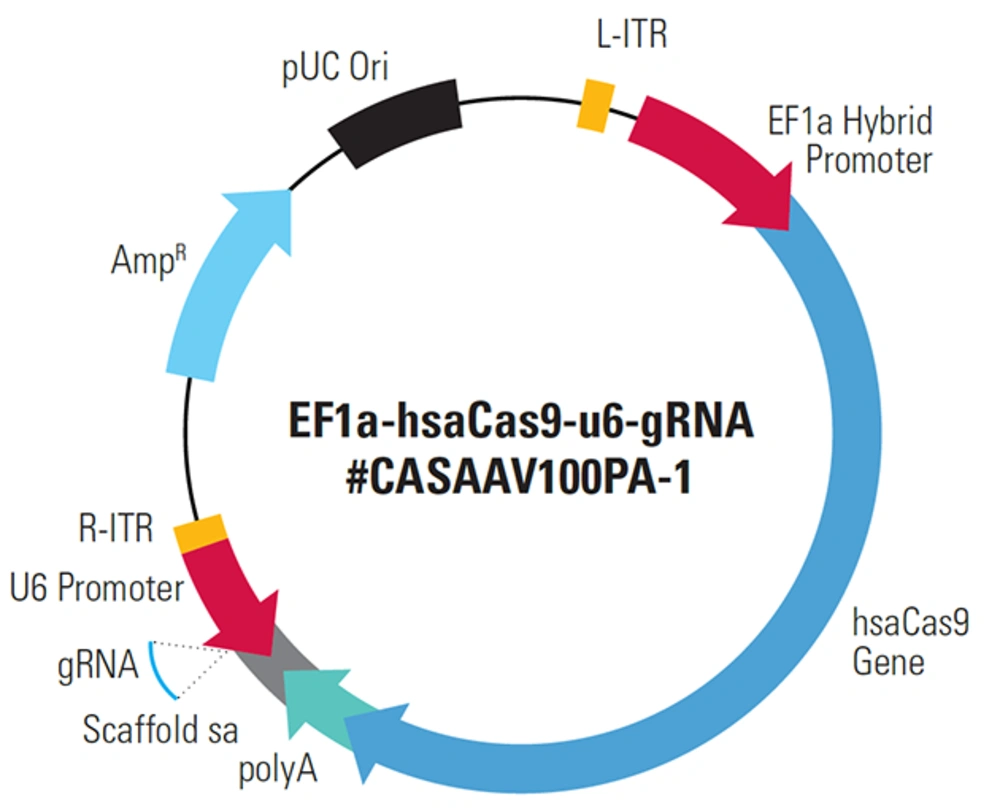
CRISPR software was used to design the sgRNA sequence, and the result was CCAGGACCGCGCCTTCCCACG. Using a system biosciences (SBI) kit, the plasmid vector was created. EF1a-hsaCas9-U6-gRNA (#CASAAV100PA-1) vector (Figure 1), a hybrid sgRNA and Cas9 carrier, was also employed. The sgRNA fragment was found between the promoter of the U6 gene and the poly-A region of the hsaCas9 gene.
3.2. Cell Preparation and Culture
The Iranian National Centre for Genetic and Biologic Resources provided the glioma cell line U87. U-87 is a cell line with epithelial morphology isolated from malignant gliomas from a male patient with a possible diagnosis of glioblastoma. These cells were cultured in the AmnioMAX culture medium (Gibco-America), which was incubated at 37.4°C and 5% CO2.
3.3. Transfection Process
The Lipofectamine CRISPRMAX kit was used to transfect U87 cells. The cells were cultured for this purpose in an RPMI culture medium with 15% FBS. The transfection effectiveness was increased by adding two concentrations of 4 and 8 µg/mL hexadimethrine bromide (polybrene) to each well after the cells had reached a confluency and density of 30 - 70%. After 48 hours, the wells' media were removed, and RPMI culture medium with 15% FBS and concentrations of 4 and 8 µg/mL polybrene was added. This mixture was then incubated for 30 minutes. The following step involved combining 103 cells, 50 ng of gRNA (80 and 160 µg/mL), 250 ng of Cas9 nuclease, 0.5 μL of Cas9 Plus solution, and 5 μL of Opti-MEM I medium. After that, 0.3 μL of CRISPRMAX was dissolved in 5 μL of Opti-MEM I and immediately added to the earlier formulations. A volume of 10 μL of this mixture was applied to each well containing U87 cells after they had been incubated for five to ten minutes. To prepare cell lysates, the cells were cultured for two to three days at 37°C before the culture medium was removed. The cells were then washed with 50 to 500 μL of PBS and lysed with 50 to 250 μL of lysis buffer.
3.4. Detection of Cas9 in Transfected Cells
In the transfected cells, Cas9 was found using flow cytometry. After 24-hour transfection, on average, 103 cells were put into each well for this purpose. After 72 hours of incubation, the cells were removed from the plate and centrifuged after being fixed with PBS and one unit of the trypsin enzyme. The cell pellet was washed in a 10% binding buffer solution, and centrifugation was then carried out for 15 minutes at 15000 rpm. In the end, a flow cytometer was used to conduct the reading.
3.5. Evaluation of DNA Methylation and hTERT Gene Expression
For hTERT gene methylation, the EZ-96 DNA Methylation-Gold™ kit (Zymo Research, Irvine, CA) was used according to the manufacturer's instructions. A concentration of 0.05 μL of metabisulphite-modified DNA was considered as a template for real-time PCR. According to the kit instructions, the amplification reaction was carried out in a volume of 20 μL and included 10 μL of the master solution (Master Mix), 0.4 μL of each of the forward (5’-CACGCGAAAACCTTCCTCAG-3’), and reverse (5’-GGCCTCGTCTTCTACAGGGA-3’) primers, 5 μL of metabisulphite-modified DNA, and 4.2 μL of distilled water. The eva-green real-time RT-PCR kit, Biotium (South Korea), was used for this reaction. 10 μL of SYBR Green, 2 μL of cDNA sample, 0.5 μL of forward and reverse primers (10 pmol) were mixed with 7 μL of nuclease-free water (Qiagen, Hilden, Germany). The GAPDH housekeeping gene was used as a reference, and hTERT gene expression was normalized using it. Evaluation of hTERT methylation and expression was carried out in cells transfected without gRNA, transfected with gRNA, as well as basal cells as a positive control.
3.6. Investigation of Telomerase Activity in Cells by Western Blot Technique
To perform western blot, the microtube containing cells was centrifuged at 1300 rpm for 10 minutes, and after discarding the supernatant, 100 µL of RIPA buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 140 mM NaCl) was added to the cell pellet. Following storing for 1 hour at -20°C, the falcon was placed at room temperature for 5 minutes and then centrifuged at 1300 rpm for 10 minutes at 4°C. Then the protein-containing supernatant was transferred to a new microtube. The SDS-PAGE gel in a single electrophoresis run was divided into stacking gel and separating gel. The stacking gel (acrylamide 5%) was poured on top of the separating gel, and a gel comb was inserted into the stacking gel. The protein isolated by SDS-PAGE was transferred to the PVDF membrane, which was blocked with a blocking solution (5% skim milk in 0.05% PBS-Tween 20) at -4°C for 12 hours. Then washing with PBS-Tween 20 was performed three times for 5 minutes, and it was incubated for 90 minutes at 25°C with 10 µg/mL monoclonal anti-hTERT antibody (Padza Co, Iran). The membrane was washed three times with PBS-Tween 20 for 5 minutes and exposed to 0.4 µg/mL secondary HRP antibody (Padza Co, Iran) for one hour at 25°C. After washing the membrane, the bands were detected using a gel documentation system.
3.7. Evaluation of Telomerase Activity in Cells Through Flow Cytometry
Telomerase protein levels were assessed using flow cytometry. The telomerase levels were assessed in cells transfected with gRNA, cells transfected without gRNA, and basal cells acting as a positive control using the same technique. For this purpose, an average of 103 cells were placed in each well in a 24-well plate. After incubating for 24 hours, the cell sediment was obtained using the cell passage method. A volume of 250 µL of a solution containing propidium iodide, according to the instructions of the Biolegend kit and Triton X100, was added to the resulting cell sediments. Cells were incubated in darkness for about an hour. The cells were then read immediately using a flow cytometry device, and telomerase level was obtained by comparison with the control group.
3.8. Evaluation of hTERT Gene Promoter Methylation
Gene methylation was evaluated using an high-resolution melting (HRM) assay. A 40-cycle-replication reaction was performed. For this purpose, enzyme activation (hot start) was performed at 95°C. The cycle began with an initial denaturation and holding period at 95°C for 10 seconds, followed by 20 seconds of annealing at 57°C. It concluded with a 25-second extension at 72 degrees. The temperature was also conducive to light absorption. Following these procedures, a melting step was carried out at a temperature range of 55 to 95°C to separate the DNA bands impacted by metabisulfite using HRM. The forward and reverse primers were in the following order: 5’-GCAATGCGTCCTCGGGTTC-3’ and 5’-CGGAGAGAGGTCGAATCGG-3’, respectively. The methylation percentage was then calculated from the ratio of heights of a cytosine peak (methylated signal) and the sum of cytosine and thymine peaks (methylated and unmethylated signal) for each cytosine in a CpG dinucleotide.
3.9. Evaluation of Apoptosis
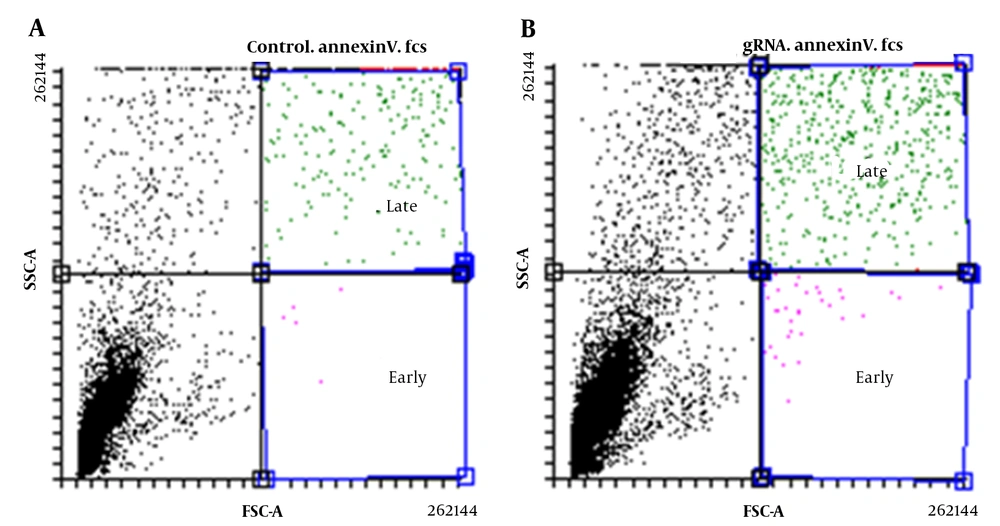
Flow cytometry was used to evaluate the apoptosis rate of U87 cells. In each well of a 24-well plate, 103 cells were cultured for this purpose, and after 72 hours, the level of apoptosis was determined. The cells were taken off the plate and centrifuged after being cleaned with PBS and one unit of the trypsin enzyme. The cell pellet was then centrifuged at 15000 rpm for 15 minutes after being rinsed with a 10% binding buffer solution. A volume of 5 μL of the Annexin V-FITC solution was incubated for 15 minutes at room temperature. Then they were prepared using 5 μL of propidium iodide and counted by flow cytometer. The results were analyzed using FlowJo software.
3.10. Statistical Analysis of Data
All experiments were carried out three times, and the results were computed as the mean ± standard deviation. The SPSS V.22 software was used for data analysis. The one-way ANOVA approach was used to compare the treatment and control groups, taking into account the normal distribution of the data, and the P-value was obtained. The threshold for significance was set at P < 0.05. A post hoc test was regarded as Tukey's test.
4. Results
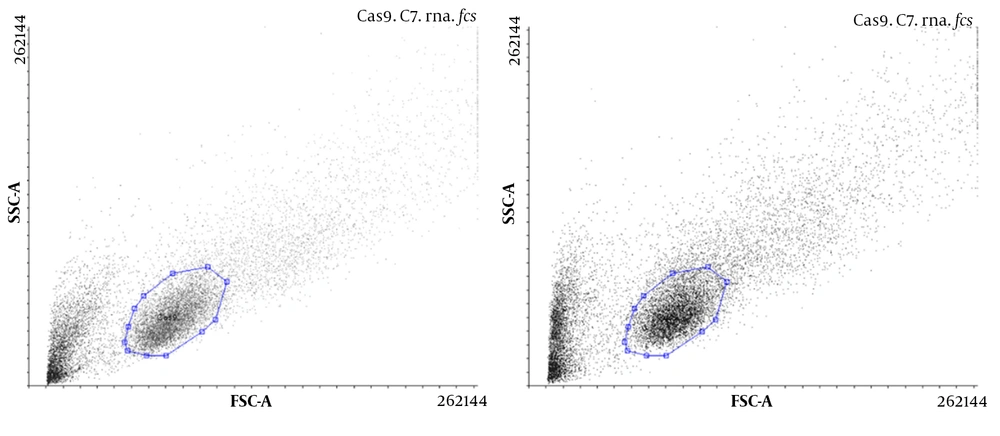
4.1. Confirmation of the Presence of Cas9 Using Flow Cytometry
The presence of Cas9 was evaluated, and the transfection efficiency was compared with and without gRNA in U87 cells using flow cytometry. The results showed that Cas9 was present in both types of transfected cells, but in the presence of Cas9, gRNA transfection was more efficient (Figure 2).
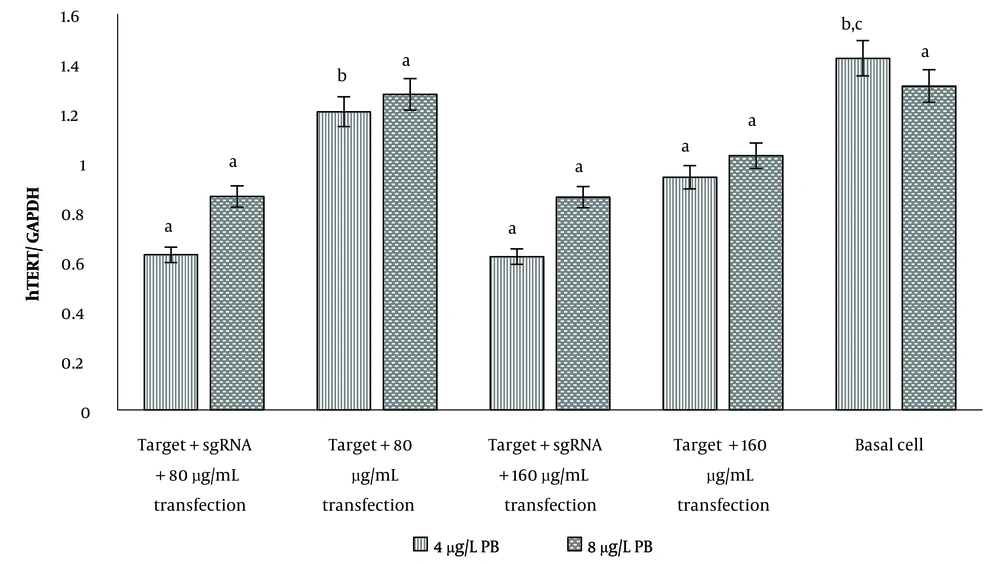
4.2. Evaluation of hTERT Gene Expression in U87 Cells
The hTERT gene expression in U87 cells showed that only at a transfection concentration of 80 μg/mL of gRNA, expression of this gene was significantly reduced compared to the case without gRNA. At the transfection concentration of 160 μg/mL of gRNA, no difference was observed between gene expression in the presence and absence of gRNA (Figure 3). In both transfection concentrations, hTERT expression showed a significant decrease compared to basal cells (P < 0.05). In transfected cells with 8 μg/mL polybrene, there was no significant difference in hTERT gene expression in the studied groups, including gRNA-transfected cells, non-transfected cells, and basal cells (Figure 3).
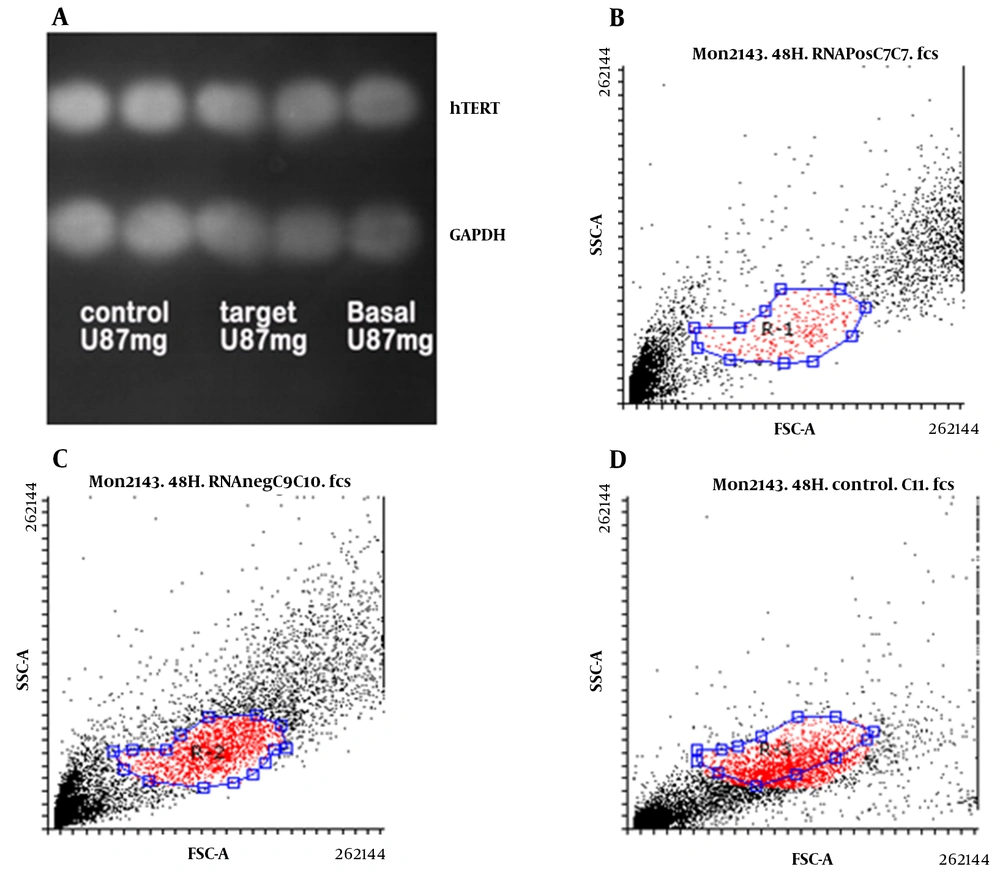
4.3. Evaluation of hTERT Protein Level
Using the Western blotting method, hTERT protein level was measured in U87 cells transfected with and without gRNA as well as basal cells relative to GAPDH protein. As shown in Figure 4A, no significant difference was observed in the amount of hTERT protein in U87 cells in the studied cells. However, examination of hTERT protein levels in U87 cells using flow cytometry revealed that this protein was higher in the basal cells (Figure 4B) and transfected cells without gRNA (Figure 4C) than in the transfected cells with gRNA (Figure 4D).
4.4. Evaluation of hTERT Gene Methylation
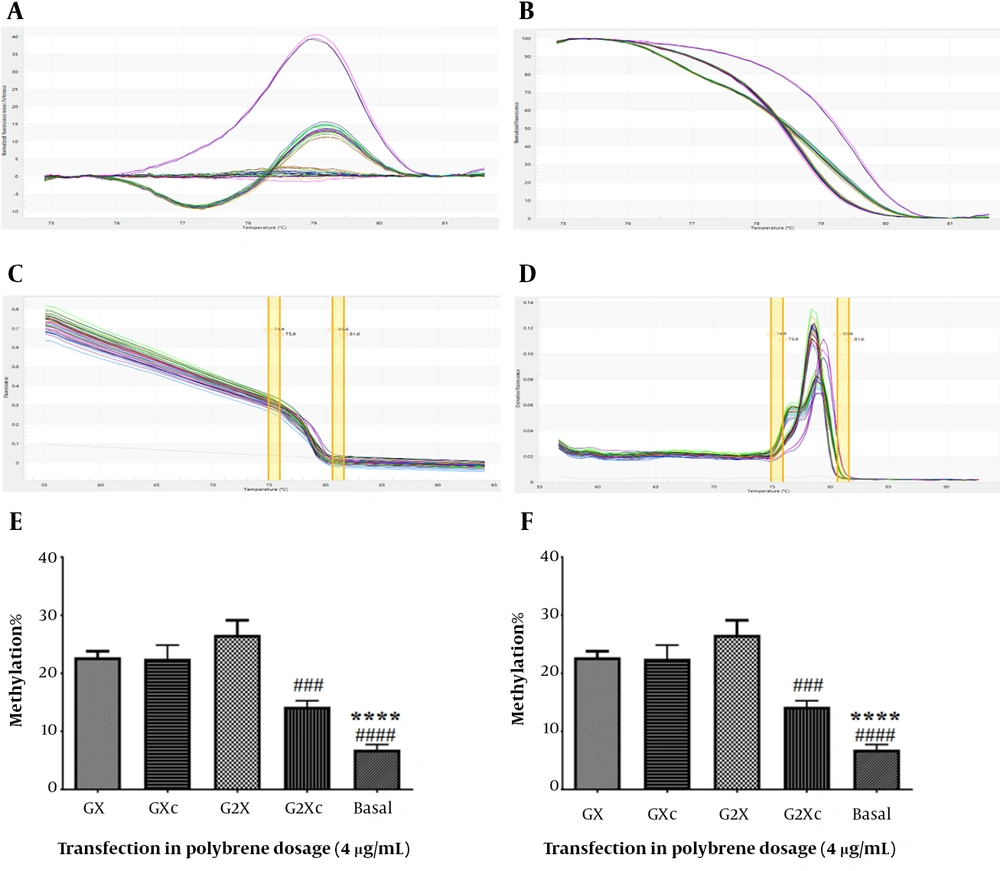
hTERT gene methylation was assessed using the HRM method (Figure 5A-D). In U87 cells at a concentration of 4 μg/mL polybrene, it was shown that only at the transfection concentration of 160 μg/mL methylation of the hTERT promoter in the presence of gRNA was significantly increased compared to its absence (Figure 5E). Compared with basal cells, both 80 and 160 μg/mL transfection concentrations significantly increased hTERT promoter methylation (P < 0.05). At a concentration of 8 μg/mL polybrene, both 80 and 160 transfection concentrations significantly increased the methylation of the hTERT promoter in the presence of gRNA relative to its absence in U87 cells (Figure 5F). Also, a significant increase in hTERT promoter methylation was observed in both 80 and 160 μg/mL transfection concentrations compared to basal cells (P < 0.05).
Evaluation of hTERT gene methylation. A, difference melting curve; B, normalized melting curve; C, component melting curve; D, derivative melting curve; E, hTERT gene promoter methylation in transfected U87 cells in the presence of 4 μg/mL polyburn; F, methylation of the hTERT promoter in transfected U87 cells in the presence of 8 μg/mL polybrene. GX: Target + sgRNA + 80 μg/mL transfection, GXc: Control + 80 μg/mL transfection, G2X: Target + sgRNA + 160 μg/mL transfection, G2Xc: Control + 160 μg/mL transfection. **: Significant difference with GX group with P < 0. 01, ****: Significant difference with GX group with P < 0.0001, #: Significant difference with G2X with P < 0.05, ###: Significant difference with G2X with P <0.001, ####: Significant difference with G2X with P < 0.0001
4.5. Evaluation of Apoptosis Rate in U87 Cells
Flow cytometry was used to assess the apoptosis rate in transfected U87 cells, and the results showed both early and late apoptosis rates were higher in transfected cells with gRNA compared to transfected cells without gRNA (Figure 6). In fact, in the absence of gRNA, the amount of early and late apoptotic cells was estimated to be 0.06% and 7.01%, respectively, while in the presence of gRNA, these values reached 0.27 and 11.37%, respectively (P < 0.05).
5. Discussion
The results of the present study indicated that in U87 cells, only at a concentration of 4 μg/mL polybrene and 80 μg/mL transfection, there was a significant difference in the expression of hTERT compared to transfection without gRNA and basal cells. Evaluation of hTERT promoter methylation also showed that in U87 cells at concentrations of 4 and 8 μg/mL polybrene and 80 and 160 μg/mL transfection, there was a significant increase in hTERT promoter methylation compared to basal cells. The level of hTERT protein in U87 cells showed an increase only in flow cytometry results. Nevertheless, Western blotting did not reveal any appreciable variations in the levels of hTERT protein in the cells. Finally, it was shown that transfection with gRNA in U87 cells increases the induction of apoptosis compared to transfection without gRNA. The present study's findings generally demonstrated that the designed CRISPR/Cas9 system could kill U87 glioma cancer cells by decreasing the expression of hTERT at the RNA and protein levels and by increasing the methylation of its promoter.
Cancer occurs when genomic instability accumulates in normal cells, and the cells reach limitless proliferation capacity. Cancer cells can delay the effects of aging by activating telomerase or using other mechanisms to prolong the length of their telomeres. In 1994, it was reported that telomerase controls 90% of malignant tumors, which is essential for the development of cancer and other diseases (14, 15). Although repetitive mutations in the hTERT promoter play a significant role in activating hTERT in malignancies, there are other tumor types (such as prostate and breast cancer) where these mutations are less common (16). Because of this, the importance of epigenetic mechanisms in regulating hTERT differs depending on the kind of cancer, and many studies have shown different results in terms of the impact of hTERT promoter methylation on hTERT regulation. Promoter methylation and gene silencing usually occur together. However, many investigations have shown that methylation of certain regions within the hTERT promoter, particularly upstream of the main promoter, is associated with gene activation (14). The specific mechanisms by which the pattern of hTERT promoter methylation results in hTERT activation are still being investigated. Recently, it has been shown that hTERT promoter methylation may play a role in activating the expression of hTERT (17).
There are various possibilities as to how hTERT activation can result from hTERT promoter methylation. Based on the prevention of suppressor elements binding in the repressor region, the first hypothesis is a DNA methylation-related mechanism. If the hTERT promoter is hypomethylated (or non-methylated), transcription inhibitors bind to the promoter and block transcription machines. If hTERT is methylated, though, it prevents this binding and makes it possible for the promoter to be turned on by proper transcription factors. Interesting observations have revealed that the upstream region of the primary promoter of hTERT is frequently hypermethylated, thus preventing the key stimulus of gene expression from accessing the promoter. Another explanation is more complicated and involves chromosome structural alterations and DNA methylation. By altering the structure of chromatin and the impact of DNA exposure on transcription factor binding, DNA methylation can influence gene expression (18). Histone modifications and DNA methylation are frequently linked, and the latter may regulate how transcription factors can access the promoter. Specific conformational changes caused by hTERT promoter methylation may result in different levels of uptake and binding of factors that may cause hTERT overexpression in cancer (19). Histone acetylation and methylation are two post-translational histone changes that can influence chromatin density and, consequently, gene expression (20).
Malignant blood, prostate, urothelium, brain, and colon tumors, among other types of cancer, have all shown a high frequency of hypermethylation in the region upstream of the major hTERT promoter. More surprisingly, hTERT promoter methylation is linked to hTERT overexpression even in melanoma, where hTERT promoter alterations were originally discovered, and a mechanism for hTERT activation was considered (21). Although this tumor-specific signature is found in a wide variety of tumor types, more work needs to be done to translate these findings to clinical settings. Pediatric glioma possesses a methylated area in the hTERT promoter that may serve as a potential biomarker for tumor survival and progression (14). Malignant tumors have hypermethylation in this region, known as THOR, while healthy tissues and stem cells have hypomethylation. Prostate cancer research has focused on THOR and has demonstrated its potential as a marker with diagnostic and prognostic properties (17).
Gigek et al. (22) examined the methylation and expression of hTERT in gastric cancer and demonstrated that hTERT was not expressed in healthy individuals, while in 80% of tumor tissues, this gene was expressed. In contrast, the hypermethylation of the hTERT promoter in tumor samples was significantly higher than in healthy samples. However, they reported that there was no significant relationship between promoter hypermethylation and hTERT expression (22). Bougel et al. (23) reported that in the cerebrospinal fluid of people with metastatic leptomeningeal cancer, approximately 1% of hTERT was methylated, and the rest was non-methylated and that the methylation of the hTERT promoter can be used as a diagnostic marker. Modification of methylation and expression of hTERT has been reported in different types of cancers (24-26), and most stated that with increasing methylation, the expression of hTERT increases. However, in the present study, it was shown that increased methylation was associated with the downregulation of hTERT. In this respect, one can say that the type of cancer studied can affect the observed mechanisms.
This work exploited the activation of hTERT promoter methylation as an epigenetic alteration to develop a treatment for glioma malignancy. Most epigenetic modifications, in contrast to gene mutations, may be reversed or prevented. In the treatment or prevention of cancer, it has become increasingly popular to restore abnormal epigenetic events in neoplastic cells (27). Before the development of CRISPR technology, scientists used a variety of instruments that could separate DNA double strands to modify the genome in various ways. As an example, four different subcategories of DNA-binding nucleases, such as Cas9 nuclease, the most modern meganucleases, zinc finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALEN), were considered. Every known nuclease has its restrictions. Zinc finger nucleases contain distinct DNA-binding domains as well as the Fok1 endonuclease, a generic cleavage domain. The main difficulty also lies in the design and building of ZNFs (28). Low binding to and separation from the target sequence is another drawback of this method, which reduces its effectiveness (29). We used the potent CRISPR technology in this study because it was a much cheaper and simpler way to make various changes to the genome.
In a prior investigation, the authors used the A-375 cell line connected to malignant melanoma of pseudoepithelial origin to analyze the methylation alterations of the major promoter of the hTERT gene and the amount of its expression. They used dCas9, a modified version of Cas9, and DNMT3A, a similar protein. The outcomes demonstrated that the developed CRISPR/Cas9 system decreases hTERT expression and telomerase, and thus it can prevent melanoma cells’ growth (30).
The PTEN gene was chosen by Moses et al. (31) as the target of the CRISPR/Cas9 system in the SK-MEL-28 melanoma cell line, and it was demonstrated that this repair system is capable of upregulating PTEN and reducing cancer cell migration. The main reason for their observations, as they stated, was decreased signaling of oncogenic pathways such as AKT, mTOR, and MAPK. In a related study, Moses et al. (31) reported that the CRISPR/Cas9 gene modification in the melanoma cell line resulted in an upregulation of PTEN. Their research used the PI3K/mTOR pathway's inhibition as the repair system's mechanism of action. In relation to glioma, studies have been used to use the CRISPR/Cas9 gene editing system to inhibit cancer cells, but according to the author's search in the databases, none of them was on the hTERT gene. Huang et al. (32) used the CRISPR/Cas9 system to target exon 17 of the EGFR gene in glioma cells, thereby activating the inhibitory modification of UBXN1 and inhibiting tumor necrosis factor in these cells. Using the CRISPR/Cas9 system, Morimoto et al. (33) inhibited the TIM3 gene in natural killer cells, thereby inhibiting the growth of glioma cells. Moure et al. (34) used the CRISPR/Cas9 editing system to knock out the IDH1 and induce CpG demethylation in glioma models. Therefore, it seems that the CRISPR/Cas9 gene editing system can be well used to inhibit the growth of cancer cells, especially by targeting the methylation status of genes, and in various studies, as mentioned, this system has been used to modify the methylation and expression of genes involved in glioma. Target genes and methylation and demyelination status can differ depending on the type of cancer's target mechanisms. What was observed in the present study was that the CRISPR/Cas9 system in glioma can downregulate this gene by increasing the promoter methylation of the hTERT gene, thereby killing cancer cells.
5.1. Conclusions
Since the development of genomic editing tools, CRISPR/Cas9 technology has revolutionized biology by facilitating the modification of the genomes, transcriptomes, and epigenomes of different organisms. Cancer research could be revolutionized by CRISPR/Cas9, but this has not yet been completely realized. This technique was employed in this study for methylation, and the outcomes showed how well this technology led to apoptosis in U87 cells. In fact, the treatment of glioma produced positive results thanks to the employment of CRISPR/Cas9 as a therapeutic option and the hTERT gene as a therapy target. In the future, CRISPR technology may target various genes in many cancer cells. CRISPR can spot deadly cancer interactions and help find novel therapeutic targets by combining genomic and epigenetic data from cancer cell lines.