1. Background
Multi-drug resistant Escherichia coli are opportunistic pathogens causing hospital-acquired infections worldwide. These infections include catheter-associated urinary tract infections, ventilator-associated pneumonia, surgical wound infections, and bacteraemia. They often carry resistance genes to antibiotics, such as β-lactams and fluroquinolone, that are commonly used for treatment. Genes encoding extended-spectrum β-lactamases (ESBLs) are often found on mobile genetic elements (MGEs) and are harbored within transposons or insertion sequences, thereby facilitating their spread to other strains. The most prevalent and dominant ESBL gene found in Enterobacteriaceae isolated from humans and food-producing animals is blaCTX-M-15 (1). Recently, a major concern has been the resistance to colistin, a polymixin, one of the last antibiotics in use after others failed. Colistin resistance gene mcr, which currently has ten variants, is usually found on plasmids of various incompatibility groups (IncX4, IncI2, and IncHI2) and often coexists with ESBLs (2, 3). In addition to ESBL genes, macrolide, tetracycline, aminoglycoside, fluoroquinolone, and carbapenem resistance genes can also coexist in a colistin-resistant isolate, limiting treatment options for hospitalized patients.
Plasmid (mcr)- and chromosomal-mediated colistin resistance involve mutations in genes encoding enzymes that are associated with outer membrane modification of LPS by encoding a phosphoethanolamine transferase that catalyzes the addition of a phosphoethanolamine moiety to lipid A (3, 4), such as the pmrC and pmrE and the pmrHFIJKLM operon (4). Previous studies on E. coli have revealed that mutations in the sensor histidine kinase pmrB are an important mechanism of colistin resistance, leading to the constitutive production of the enzymes ArnT and EptA that add a positive charge (4-amino-4-deoxy-L-arabinose and phosphoethanolamine, respectively) to the phosphate groups of lipid A and reducing the affinity of colistin to bind to lipid A (3, 4).
To plan effective treatment guidelines, it is crucial to understand the mechanisms of resistance and epidemiology of multidrug-resistant (MDR) E. coli in both the community and hospitals. Given the burden of diseases caused by E. coli and its significant public health concern, hospitals should continuously monitor their antimicrobial treatment efficacy. Whole-genome sequencing (WGS)-based in silico approaches are valuable tools in gene analysis of outbreak strains that offer detailed epidemiological investigation and tracing of pathogens (5). In this study, we used WGS to characterize E. coli C91 (ST38), an extensively drug-resistant clinical outbreak strain isolated from patient zero in the intensive care unit (ICU) of one of the largest hospitals in Kuwait, with the intention of successfully treating the patients and containing its spread.
2. Methods
2.1. Sample Collection
A clinical E. coli isolate C91 was isolated from a post-surgical wound of a 53-year-old male admitted to ward 8/ICU (26/11/2016) and was initially identified by VITEK 2 ID system (bioMérieux, Marcy-l’Etoile, France). This patient was named patient zero.
2.2. Antibiotic Sensitivity Testing
Antimicrobial sensitivity testing was carried out according to the Clinical and Laboratory Standards Institute (2020) (6). The minimum inhibitory concentrations (MICs) were determined for aminoglycosides, chloramphenicol, tetracycline, β-lactams, including carbapenems, and in combination with β-lactam inhibitors, ciprofloxacin, erythromycin, trimethoprim, gentamycin, and colistin. The MIC (μg/mL) against a panel of antibiotics were determined using E test (bioMérieux, Marcy-l’Etoile, France). For colistin, the agar dilution method was used (6).
2.3. Whole-Genome Sequencing Analyses
Genomic deoxyribonucleic acid (DNA) was extracted using QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany) and quantified by the NanoDrop-800 spectrophotometer (Thermo Fisher Scientific, Wilmington, NC, USA) according to the manufacturer’s instructions. The WGS was performed by MicrobesNG, University of Birmingham, UK (https://microbesng.uk) using the Illumina MiSeq® sequencer platform. The reads were trimmed using Trimmomatic, and the quality was assessed by MicrobesNG’s in-house scripts combined with the following software packages: SAMtools (Sequence, Alignment/Map), Bedtools, and bwa-mem (Burrows-Wheeler Aligner). All statistics are based on contigs of size ≥ 500 bp unless otherwise noted. The trimmed data were assembled using the SPAdes algorithm assembler (version: 3.7.1); this de novo assembly of the quality-controlled reads was assembled to create a draft genome sequence, and variant calling was performed using VarScan. An automated annotation was performed using Prokka (version 1.13.3). The WGS of the isolate was submitted to Genbank Accession: SAMN10105215, ID: 10105215 (sample name: Escherichia coli strain Kuwait C-91).
2.4. In Silico Molecular Analysis
For in silico WGS analysis, the assembled sequences were uploaded onto the Center for Genomic Epidemiology to identify the following: ResFinder v4.3.3, ResFinderFG 2.0, KmerResistance 2.2 (7), PathogenFinder1.1 (8), VirulenceFinder 2.0 (9-11), multilocus sequence typing 2.0 (12), PlasmidFinder 2.1 (13), MGE v1.0.3 (14), SerotypeFinder 2.0 (15), FimTyper 1.0 (16), and 2.1 (13), CHTyper 1.0 (17), CARD 2020 annotation (18), Pfam (InterPro 95.0), VirSorter2 version 2.2.4 (https://u.osu.edu/viruslab/). The presence of insertion sequences was confirmed using ISFinder (19). Proksee CGView.js server was used for genome assembly, annotation, and visualization and provided a complete genome CGView/Proksee map JSON file (20).
2.5. Detection of Phages from WGS
Phaster tool (21) was used to identify prophage sequences. This tool classifies the phages into three classes (intact, questionable, and incomplete) based on their completeness (phage score). Additionally, by using the Proksee server, phages were identified with the VirSorter2 2.2.4 tool and were screened for antimicrobial resistance genes using the basic local alignment search tool (BLAST).
3. Results
3.1. Description of the Isolate and Mapping Summary
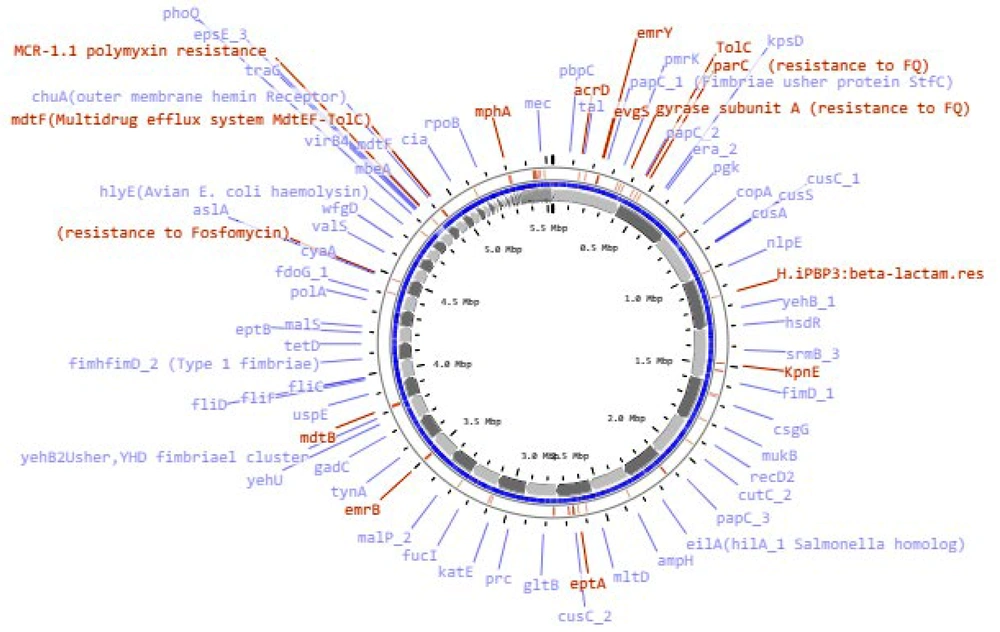
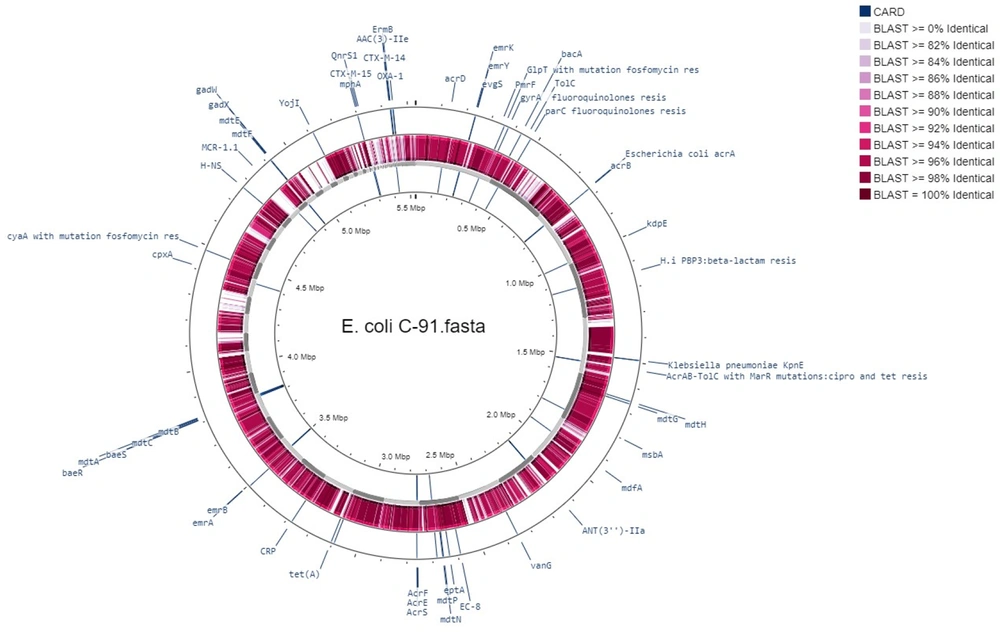
The bacterial strain C91 was identified as E. coli O99 H30 ST38 according to two different schemes, Warwick and Pasteur Institute (Appendix 1). The draft genome was annotated using RAST (Table 1) and revealed a linear chromosome consisting of 5 532 235 base pairs, with 4 964 coding sequences, 87 transfer RNA (tRNA) genes, and several proteins with functional assignments. The genome was assembled using the SPAdes assembler (version: 3.7.1) from trimmed data, producing an N50 quality value of 181 117 and a L50 of 11, with an N75 of 97 722 and L75 of 20. The sample had a mapping rate of 76.81% against the reference genome (without Ns), with an average depth of 76.96X and over 90.93% coverage of more than 1X, a result that falls within the normal range. The genome mapping of antimicrobial resistance and virulence factors is shown in Figure 1, and the comparison of E. coli C91 to E. coli K12 MG1655 (GenBank: U00096.2) using NCBI and Proksee software is presented in Figure 2.
| E. coli C91 | Values |
|---|---|
| Genome size (bp) | 5 532 235 |
| Total length of the genes (bp) | 4 529 853 |
| GC content % | 51.45 |
| Number of genes | 4 964 |
| % of genome (genes) | 86.67 |
| Gene average length (bp) | 913 |
| Gene internal length | 696 465 |
| Gene internal GC content | 43.87 |
| % of genome (internal) | 13.33 |
| Average depth | 76.96X |
| Contigs | 183 |
| Largest contig | 399 443 |
| Genome coverage | 90.93% |
| GC% | 50.42 |
| N50 | 181 117 |
| N75 | 97 722 |
| N90 | 66 020 |
| L50 | 11 |
| L75 | 20 |
| sRNAs | 78 |
| tRNAs | 87 |
3.2. Plasmids and MGEs
Five plasmids IncY, IncI2(Delta), IncFIC(FII), IncI1-I(Alpha), and IncFIBIncF and 17 MGEs, including Tn7, were detected harboring antibiotic resistance genes. Their locations are shown in Table 2.
| Plasmid | Contig | Position in Contig | Coverage % | Identity % | Accession No. | Resistance Genes/Phage |
|---|---|---|---|---|---|---|
| IncY | NODE_22_length_94041_cov_7.78437 | 24344..25108 | 100 | 99.74 | K02380 | Circular phage/ PHAGE_Salmon_SJ46_NC_031129(89) |
| IncI2(Delta) | NODE_29_length_60972_cov_14.1908 | 4841..5156 | 100 | 98.42 | AP002527 | mcr-1.1 |
| IncFIC(FII) | NODE_32_length_48168_cov_7.4304 | 2747..3243 | 99.4 | 94 | AP001918 | - |
| IncI1-I(Alpha) | NODE_33_length_45415_cov_9.87703 | 15412..15553 | 100 | 99.3 | AP005147 | - |
| IncFIB | NODE_53_length_5159_cov_9.67925 | 2548..3229 | 100 | 97.65 | AP001918; CP053724 | - |
3.3. Antibiotic Sensitivity Testing and Resistance Genes
E. coli C91 was resistant to all antibiotics tested, including aminoglycosides, chloramphenicol, tetracycline, β-lactams (both alone and in combination with β-lactam inhibitors), ciprofloxacin, erythromycin, trimethoprim, gentamycin, and colistin. The MIC for these antibiotics was greater than 32 mg/L. It also demonstrated intermediate resistance to imipenem and meropenem with an MIC of 4 mg/L. The analysis of the genome of E. coli C91 revealed the presence of 200 antibiotic-resistance genes, including efflux pump complexes and antibiotic target protection proteins, as confirmed by CARD annotations. The genome analysis also revealed the presence of mcr-1, blaCTX-M-14, blaCTX-M-15, and blaOXA-1 genes, but not blaNDM or blaVIM. The bacterium was observed to have acquired resistance genes, including aac(3)-IIa, aac(6')Ib-cr, aadA1, qnrS1, catB4, tetA, mphA, ermB, and dfrA1, as shown in Table 3 (and Appendix 2) and Figure 1. Point mutations were also detected in chromosomal resistance genes, including gyrA, parC, and pmrB, leading to changes in amino acids, as shown in Table 4. The results of some of these mutational modifications are not clear.
| Gene | Phenotype | Position in Contig/MGE, Plasmid | Coverage % | Identity | Accession |
|---|---|---|---|---|---|
| D-alanine--D-alanine ligase van_ligase | Cycloserine | NODE_4_length_294518_cov_34.7303_119741_118821 | 100 | 99.02 | KF628564.1 |
| D-alanyl-D-alanine carboxypeptidase, none enzyme β_lactamresistance | Penicillin | NODE_7_length_249411_cov_31.6868_90740_91963 | 99 | 99.75 | BDB50754.1 |
| ant(3'')-Ia, (aadA1) | Spectinomycin, streptomycin | NODE_8_length_222074_cov_39.2814_40651_39863/Tn7 | 100 | 100 | JQ480156 |
| dfrA1 | Trimethoprim | NODE_8_length_222074_cov_39.2814_41801_41328/Tn7 | 100 | 100 | X00926 |
| Multidrug resistance protein MdtL | NODE_8_length_222074_cov_39.2814_62182_61007 | 100 | 100 | WP_000086009.1 | |
| Multidrug efflux MFS transporter EmrD | NODE_8_length_222074_cov_39.2814_102857_101673 | 100 | 99 | WP_097336506.1 | |
| van_ligase | D-cycloserine | NODE_9_length_221455_cov_34.9789_34311_35405 | 100 | 99.02 | KF628791.1 |
| β-lactamase | Piperacillin | NODE_10_length_210474_cov_36.1065_48903_50069 | 100 | 99.83 | KU607300.1 |
| emrE (SMR protein family) | Ethidium multidrug resistance | NODE_12_length_170122_cov_27.8913/ISEc46 | |||
| tet(A) | Tetracycline, oxytetracycline, doxycycline; minocycline | NODE_13_length_161358_cov_37.5202_2716_3915/Tn5403 | 100; 100 | 100; 99.85 | AJ517790; JX009293.1; GQ343144.1 |
| sitABCD | Hydrogen peroxide | NODE_16_length_138200_cov_28.3328_4692_1243 | 99.59 | 97.48 | AY598030 |
| Multidrug efflux system MdtABC-TolC | NODE_17_length_131052_cov_29.4018_123533_115966 | 100 | 100 | CP128875.1 | |
| mcr-1.1 | Polymyxin, colistin | NODE_29_length_60972_cov_14.1908_47111_45486/ Incl2(Delta) | 100 | 100 | KP347127; OM179755.1 |
| qnrS1 | Ciprofloxacin | NODE_43_length_11726_cov_19.4543_6035_5379/ISKpn19 | 100 | 100 | AB187515 |
| mph(A) (Macrolide phosphotransferase) | Azithromycin, telithromycin, erythromycin, spiramycin | NODE_43_length_11726_cov_19.4543_197_1102/ISKpn19 | 100 | 100 | D16251 |
| blaCTX-M-15 (Class A) | Ticarcillin, aztreonam, ampicillin, amoxicillin, piperacillin, ceftazidime, cefotaxime, ceftriaxone, cefepime | NODE_43_length_11726_cov_19.4543_11551_10676/ISKpn19 | 100 | 100 | AY044436, GQ343005.1 |
| blaCTX-M-14; (Class A, blaCTX-M-14a-like) | Ticarcillin, aztreonam, ampicillin, amoxicillin, piperacillin, ceftazidime, cefotaxime, ceftriaxone, cefepime | NODE_65_length_3010_cov_7.26882_2841_1966/IS102 | 100; 100 | 100; 99.89 | AF252622; KU544013.1 |
| aminoglycoside N(3')-acetyltransferase III gene; aac(3)-IIe | Gentamicin | NODE_66_length_2854_cov_39.711_171_1031/ISKpn19 | 100 | 100 | GQ343134.1; CP125071; HCQ1792082.1 |
| aac(3)-IIa | Gentamycin, tobramycin | NODE_66_length_2854_cov_39.711_171_1031//ISKpn19 | 100 | 100 | CP023555 |
| erm(B) | Macrolide, lincosamide, streptogramin, quinupristin/dalfopristin | NODE_67_length_2837_cov_7.75646_420_1157 | 100; 100 | 99.73; 99.86 | JN899585; CP082057 |
| aac(6')-Ib-cr | Fluoroquinolone, ciprofloxacin, dibekacin, sisomicin, netilmicin, amikacin, tobramycin | NODE_70_length_2440_cov_46.4838_174_773 | 100 | 100 | DQ303918; GQ342986.1 |
| blaOXA-1 | Carbenicillin, ampicillin, amoxicillin, piperacillin, cefepime, ampicillin+clavulanic acid, amoxicillin+clavulanic acid, piperacillin+tazobactam | NODE_70_length_2440_cov_46.4838_859_1734 | 100 | 100 | HQ170510; MN340011.1 |
| catB3 | Chloramphenicol | NODE_70_length_2440_cov_46.4838_1872_2420 | 70 | 100 | U13889; AJ009818; KU544029.1 |
| Mutation | Nucleotide Change | Amino Acid Change | PMID | Notes |
|---|---|---|---|---|
| gyrA p.S83L | TCG → TTG | S → L | 8891148, 2168148, 12654733, 12654733 | |
| gyrA p.D87N | GAC → AAC | D → N | 12654733, 12654733, 12654733, 22878251, 12654733, 1850972 | D87G or D87Y confer resistance to nalidixic acid only, if occurring alone. Unknown phenotype if D87H occurs alone |
| gyrA:p.D678E | GAC → GAA | D → E | Phenotype not found in database | Unknown phenotype |
| parE p.S458A | TCG → GCG | S → A | 14506034, 28598203 | Unknown phenotype if S458T or S458A occurs alone. Nalidixic acid and ciprofloxacin resistance when associated with gyrA mutations |
| parC p.S57T | AGC → ACC | S → T | 14510643 | Unknown phenotype if S57T occurs alone. Nalidixic acid and ciprofloxacin resistance when associated with gyrA |
| parC p.S80I | AGC → ATC | S → I | 8851598, 8851598, 21856834-20638608, 8524852, 25631675, 25631675, 25631675 | Unknown phenotype if each mutation occurs alone. Nalidixic acid and ciprofloxacin resistance when associated with gyrA mutations |
| parC:p.E62K | GAA → AAG | E → K | Phenotype not found in database | Unknown phenotype |
| parC:p.D475E | GAT → GAA | D → E | Phenotype not found in database | Unknown phenotype |
| parC:p.K200N | AAA → AAT | K → N | Phenotype not found in database | Unknown phenotype |
| parC:p.L344R | CTG → CGG | L → R | Phenotype not found in database | Unknown phenotype |
| parC:p.D197E | GAC → GAG | D → E | Phenotype not found in database | Unknown phenotype |
| parC:p.D309E | GAT → GAG | D → E | Phenotype not found in database | Unknown phenotype |
| ampC promoter:p.R24 | CGA → TGA | R → * | Phenotype not found in database | Unknown phenotype |
| pmrB:p.H2R | CAT → CGT | H → R | Phenotype not found in database | Unknown phenotype |
| pmrB:p.D283G | GAC → GGC | D → G | Phenotype not found in database | Unknown phenotype |
| pmrB:p.Y315F | TAT → TTT | Y → F | Phenotype not found in database | Unknown phenotype |
3.4. Virulence Factors
This isolate has an abundance of virulence factors shown in Table 5 and Figure 1, including fimbriae, biofilm, and capsule formation genes.
| Virulence Factor | Identity | Query/Template Length | Contig | Position in Contig | Protein Function | Accession Number |
|---|---|---|---|---|---|---|
| AslA | 98.31 | 1656/1656 | NODE_24_length_92227_cov_34.925 | 37443..39098 | Contributing to the invasion of brain microvascular endothelial cells | CP022686 |
| aamR:FN554766 | 99.84 | 645/645 | NODE_2_length_314467_cov_36.7095 | 209279..209923 | Not known | |
| Air | 95.16 | 4604/4605 | NODE_8_length_222074_cov_39.2814 | 120721..125324 | Enteroaggregative immunoglobulin repeat protein | CP003034 |
| Anr | 96.24 | 213/213 | NODE_32_length_48168_cov_7.4304 | 4169..4381 | AraC negative regulator | AL391753 |
| capU | 99.91 | 1089/1089 | NODE_38_length_25756_cov_32.6998 | 7151..8239 | Hexosyltransferase homolog | CU928145 |
| chuA | 100 | 1983/1983 | NODE_30_length_56813_cov_40.9025 | 37851..39833 | Outer membrane hemin receptor | UFZU01000002 |
| Cia | 100 | 147/147 | NODE_33_length_45415_cov_9.87703 | 8729..8875 | Colicin | QMGM01000002 |
| csgA | 92.98 | 456/456 | NODE_6_length_255487_cov_30.7367 | 82846..83301 | curlin major subunit CsgA (biofilm) | CP069646 |
| eilA | 98.65 | 1698/1698 | NODE_8_length_222074_cov_39.2814 | 131902..133599 | Salmonella HilA homolog | FN554766 |
| espY2:000868321 | 94.56 | 570/570 | NODE_4_length_294518_cov_34.7303 | 145342..145911 | Not known | |
| fdeC | 92.15 | 4214/4254 | NODE_9_length_221455_cov_34.9789 | 120658..124871 | intimin-like adhesin FdeC | AP010953 |
| fimH | 100 | 489/489 | NODE_20_length_97722_cov_41.3206 | 15817..16305 | Type 1 fimbriae | NA |
| Gad | 99.1 | 1116/1401 | NODE_96_length_1120_cov_51.0514 | 1..1116 | Glutamate decarboxylase | FN554766 |
| hlyE | 98.91 | 918/918 | NODE_27_length_69431_cov_30.2889 | 62576..63493 | Avian E. coli haemolysin | ECU57430 |
| Hra | 95.01 | 741/741 | NODE_20_length_97722_cov_41.3206 | 95294..96034 | Heat-resistant agglutinin | CP040456 |
| Hra | 100 | 792/792 | NODE_2_length_314467_cov_36.7095 | 219779..220570 | Heat-resistant agglutinin | CP043942 |
| Iss | 100 | 294/294 | NODE_40_length_20447_cov_25.9281 | 19924..20217 | Increased serum survival | CP001846 |
| kpsE | 100 | 1149/1149 | NODE_2_length_314467_cov_36.7095 | 155149..156297 | Capsule polysaccharide export inner-membrane protein | AAMK02000004 |
| kpsMII_K5 | 100 | 777/777 | NODE_2_length_314467_cov_36.7095 | 141362..142138 | Polysialic acid transport protein; Group 2 capsule | MG739441 |
| neuC | 100 | 1176/1176 | NODE_2_length_314467_cov_36.7095 | 145757..146932 | Polysialic acid capsule biosynthesis protein | JJLW01000144 |
| nlpI | 99.77 | 885/885 | NODE_11_length_181117_cov_35.2668 | 107595..108479 | lipoprotein NlpI precursor | CP000243 |
| sitA | 100 | 915/915 | NODE_16_length_138200_cov_28.3328 | 3778..4692 | Iron transport protein | HG977190 |
| terC | 98.46 | 714/714 | NODE_13_length_161358_cov_37.5202 | 84643..85356 | Tellurium ion resistance protein | CP000468 |
| terC | 98.54 | 959/966 | NODE_11_length_181117_cov_35.2668 | 173664..174622 | Tellurium ion resistance protein | MG591698 |
| traJ | 98.55 | 690/690 | NODE_32_length_48168_cov_7.4304 | 34241..34930 | Protein TraJ (positive regulator of conjugal transfer operon) | AF550679 |
| traT | 100 | 777/777 | NODE_32_length_48168_cov_7.4304 | 13597..14373 | Outer membrane protein complement resistance | AAJW02000025 |
| yehA | 95.85 | 1035/1035 | NODE_17_length_131052_cov_29.4018 | 90990..92024 | Outer membrane lipoprotein, YHD fimbriae cluster | CP042934 |
| yehB | 97.5 | 2481/2481 | NODE_17_length_131052_cov_29.4018 | 88494..90974 | Usher, YHD fimbriae cluster | CP042934 |
| yehC | 96.3 | 675/675 | NODE_17_length_131052_cov_29.4018 | 87804..88478 | Chaperone, YHD fimbriae cluster | CP042934 |
| yehD | 97.24 | 543/543 | NODE_17_length_131052_cov_29.4018 | 87181..87723 | Major pilin subunit, YHD fimbriae cluster | CP042934 |
3.5. Phage Analysis
Eleven prophage regions were identified in E. coli C91, from contig 1 - 45, using the Phaster tool (Table 6 Appendix 3). Out of these regions, three are intact, seven are incomplete, and one is questionable. However, when the VirSorter2 2.2.4 tool was used in Proksee software, phages were also picked up from nodes 46-183 (Table 6). One of the intact phages is PHAGE_Salmon_SJ46_NC_031129(89)(IncY), located on NODE_22_length_94041_cov_7.78437. On NODE_12, PHAGE_Entero_SfI_NC_027339(6) (partial sequence) was detected, containing ISEc46 and emrE (ethidium multidrug resistance protein E), an SMR protein family.
| Region | Region Length (kb) | Completeness | # Total Proteins | Most Common Phage | GC % | |
|---|---|---|---|---|---|---|
| NODE_5_length_285039_cov_26.0736 | 1 | 44.1 | Intact | 54 | PHAGE_Entero_P88_NC_026014(33) | 52.82 |
| 2 | 16.3 | Questionable | 24 | PHAGE_Salmon_118970_sal3_NC_031940(4) | 50.68 | |
| NODE_12_length_170122_cov_27.8913 | 3 | 26.8 | Incomplete | 24 | PHAGE_Entero_SfI_NC_027339(6) | 45.39 |
| NODE_18_length_114548_cov_26.6919 | 4 | 26.9 | Incomplete | 21 | PHAGE_Shigel_POCJ13_NC_025434(6) | 45.95 |
| NODE_19_length_110482_cov_29.551 | 5 | 27.8 | Incomplete | 31 | PHAGE_Entero_phiP27_NC_003356(13) | 48.50 |
| NODE_22_length_94041_cov_7.78437 | 6 | 92.5 | Intact | 117 | PHAGE_Salmon_SJ46_NC_031129(89) | 48.07 |
| NODE_34_length_38724_cov_8.49753 | 7 | 9.1 | Incomplete | 14 | PHAGE_Rhodoc_RGL3_NC_016650(1) | 56.85 |
| NODE_39_length_24718_cov_29.6434 | 8 | 24.3 | Intact | 28 | PHAGE_Pseudo_phiPSA1_NC_024365(7) | 48.89 |
| NODE_40_length_20447_cov_25.9281 | 9 | 19.9 | Incomplete | 20 | PHAGE_Entero_lambda_NC_001416(19) | 56.34 |
| NODE_43_length_11726_cov_19.4543 | 10 | 8.9 | Incomplete | 11 | PHAGE_Microc_MaMV_DC_NC_029002(2) | 51.76 |
| NODE_45_length_8140_cov_11.3055 | 11 | 7.6 | Incomplete | 9 | PHAGE_Escher_RCS47_NC_042128(3) | 48.13 |
a Region: The number assigned to the region. Region length: The length of the sequence of that region (in bp). Completeness: A prediction of whether the region contains an intact or incomplete prophage. # Total proteins: The number of ORFs present in the region. Most common phage: The phage(s) with the highest number of proteins most similar to those in the region. GC %: The percentage of GC nucleotides of the region.
4. Discussion
Colistin-resistant E. coli is one of the most important nosocomial pathogens with limited treatment options. The present study characterized a multi-drug resistant clinical E. coli (C-91) isolate causing complications in the ICU of one of the largest hospitals in Kuwait. This isolate has a diverse collection of genes conferring resistance to an array of antimicrobial agents. It contains blaCTX-M-15, the most dominant ESBL (22), and blaCTX-M-14 (23), in addition to other important resistance genes, including aadA1, aac(3)-IIa, aac(6')-Ib-cr, blaOXA-1, mcr-1.1, mph(A), erm(B), catB3, qnrS1, tet(A), dfrA1, and mphA (the most common azithromycin resistance gene detected in E. coli). It encodes for resistance enzyme MPH(2')-I, which inactivates 14-membered macrolides (e.g., erythromycin, telithromycin, roxithromycin) over 16-membered macrolides (e.g.tylosin and spiramycin) (24). In this study, aac(3)-IIa, qnrS1, mph(A), and blaCTX-M-15 genes were associated with insertion sequence (IS) ISKpn19. blaCTX-M-14 was associated with IS102, tet(A), terC with Tn5403, and ant(3'')-Ia, (aadA1), dfrA1 with Tn7. In total, we identified 14 insertion sequences and transposons (Appendix 4). Insertion sequence elements can play an integral role in the transfer of these resistance genes and virulence factors in their surrounding regions (25).
Escherichia coli C91 also contains several multidrug efflux pumps, including the multidrug efflux pump mdf(A), which confers resistance to antibiotics, such as chloramphenicol, erythromycin, and fluoroquinolones (26). The present study also detected mutations in chromosomally encoded gyrA, gyrB, parC, pmrB, ampC, and cya genes causing resistance to fluoroquinolones, polymyxins, and fosfomycin. We did not detect blaNDM, blaVIM, nor blaOXA-48 in this isolate, althrough the MIC for imipenem was just below the cutoff point (MIC = 4). However, others have reported the prevalence of carbapenem resistance among Enterobacteriaceae in hospitals in Kuwait (27).
Antimicrobial resistance plasmids present in E. coli C91 comprise epidemic resistance plasmids IncFIB and IncFIC(FII), which can acquire resistance determinants and disseminate readily among Enterobacteriaceae and broad-range IncY, IncI2 (pMCR-1) carrying the mcr-1 gene. IncI1-I plasmids have been shown to propagate the resistance genes between different species (28). Therefore, this isolate has the potential to tolerate and resist conventional antibiotic therapies.
The identification of E. coli clones in the fields of taxonomy and epidemiology is predicated on a combination of O- and H- antigens. These antigens are characterized by variations in the sugars present in the O unit and the linkages between O units (29). There are currently 185 O antigens, and the O99 antigen consists of four d-rhamnose moieties in the backbone and two d-glucose moieties in the side chain. The O-antigen is synthesized and transported by an ABC transporter-dependent process and is considered an important virulence factor, offering selective advantages in specific niches. Pathogenic clones are often found to have a higher incidence of certain O antigens (29, 30).
H-antigens (flagellins) are encoded by fliC genes, with 53 different serotypes of H-antigen identified (31). The diversity of H-antigens arises from lateral gene transfer and recombination of foreign DNA, generating alleles and antigenic variation (32). FimH genes encode a type I fimbria that enables adherence and infects the epithelial urinary tract tissue expressed in uropathogenic E. coli (UPEC). FliC genes encode proteins that promote successful host colonization and are involved in interleukin-6 (IL-6) and interleukin-8 (IL-8) release. FumC genes encode a protein that catalyzes fumarate oxidation to malate during the oxidative TCA cycle under aerobic conditions. FumC is required for E. coli fitness in vivo, and a loss of FumC results in delayed growth during iron limitation (33-35).
The H30 subclone has been reported to be responsible for the clonal dissemination of ST131 E. coli (36). Therefore, it is proposed that H30 provides ST38 clones with the advantage of propagation. Since E. coli sequence type ST38 has become prominently associated with hospital- and community-acquired infections worldwide (37-39), it is crucial to identify the subclones to increase the chances of successful treatments.
In conclusion, E. coli C91 (ST38) O99 H30 is a high-risk and globally disseminated extraintestinal pathogenic (ExPEC) strain that can cause invasive infections and resist multiple antibiotic treatments. This study used WGS and in silico analysis to identify the molecular characteristics of this isolate. The obtained results showed that it contains genes encoding ESBLs that confer resistance to cephalosporins and other β-lactam antibiotics. Additionally, E. coli C91 (ST38) is resistant to macrolides, tetracyclines, aminoglycosides, and fluoroquinolones, making it extensively drug-resistant (XDR). Furthermore, it carries mcr-1 gene, which severely limits the treatment options. This isolate also encodes several virulence factors facilitating biofilm formation and adherence to tissues. Infections caused by XDR E. coli C91 (ST38) O99 H30 in the ICU might be life-threatening and require urgent treatment.