1. Context
Enzymes are biological catalysts that are an essential component of biological reactions (1). Most bacteria, yeasts, and fungi are capable of producing amylase. Aspergillus Oryzae and Rhizopus sp. are important fungi that produce amylolytic enzymes (2). Enzymes are now used in various parts of the industry (3). They are used in detergents, paper industries, textile industries, food industries, and many other industrial applications (4). Alpha-amylase also has important applications in the medical industry. According to studies, it is used to diagnose and treat acute pancreatitis and to differentiate pancreatitis from other causes of acute abdominal diseases. As many as 80% of patients with acute pancreatitis have high levels of amylase. This test is required for the identification of symptoms of pancreatic diseases, such as severe abdominal pain, fever, loss of appetite, and vomiting (5). Serum amylase levels are increased slightly in benign inflammation of the pancreas and decrease after two days, and this pattern can be used to diagnose this type of disease. This enzyme is sometimes used to treat pancreatic cancer and gallstones. Alpha-amylases have been used to reduce starch, produce beverages, such as beer, treat digestive disorders, and produce cheese from milk (6). Among the most widely used enzymes, alpha-amylase hydrolyzes the carbohydrate chain of starch at random locations to produce short oligosaccharides, maltose, and glucose (7).
The demand for amylase is increasing due to its important role in starch hydrolysis and applications of this hydrolysis process. Compared to chemical methods that require harsh conditions, such as high pressure and high temperatures, the use of microorganisms in the production of enzymes will be much more economical (8). On the other hand, the demand for high-quality products leads to new methods for improving industrial products, such as protease and amylase, which are often used in industry and medical sciences (9). Most alpha-amylases are secreted extracellularly, but some alpha-amylase are secreted intracellularly (10). Due to the prominent application of alpha-amylase, there is an urgent need to develop cost-effective techniques for the production of stable and efficient alpha-amylase (11). The immersion fermentation method is the traditional method in the production of alpha-amylase.
In general, for the industrial production of more important enzymes, the immersion fermentation method is used because indicators, such as acidity, temperature, aeration, oxygen transfer, and humidity are better controlled (12). Because of a high volume of enzyme production, it easily meets the needs of different industries. The growth and production of enzymes by microorganisms are highly dependent on the compounds and factors entering the system (13). Optimization is the use of one factor at a time so that one of the factors (response variable) changes and the other variables are kept constant (13). The aim of the present systematic review was to assess the optimized content medium to enhance alpha-amylase production from the microbial source.
2. Methods
2.1. Search Strategy
A comprehensive search of PubMed, Embase, ISI Web of Science, Scopus, and Google Scholar was done from inception to April 2022 by two researchers (B. E. and R. H.) independently. The literature search was restricted to the English language. Combinations of the following terms were used in the search strategy: ‘Alpha-amylase’ OR ‘α-amylase’ and ‘optimization’ OR ‘improvement’ AND ‘production’ AND ‘purification’. The eligible articles were used, and their reference lists were also checked for potential additional articles.
2.2. Eligibility Criteria
2.2.1. Inclusion Criteria
Studies focused on alpha-amylase production, optimization using statistical design, and other important characterizations were worthy of inclusion. Most articles had evaluated the optimization of alpha-amylase production using statistical experimental designs.
2.2.2. Exclusion Criteria
Exclusion criteria were as follows: (1) papers that only reported alpha-amylase activity; (2) articles that only focused on enzyme screening studies or did not assay alpha-amylase activity; (3) studies that lack parts about the screening of significant factors by Plackett-Burman design; (4) conference abstracts, reviews, letters, personal opinions, and book chapters.
2.3. Data Extraction
Two researchers (B. E. and R. H.), according to the above-mentioned inclusion and exclusion criteria, independently extracted and recorded the following information from eligible papers: First author’s name, publication year, microbial species, alpha-amylase activity, growth conditions, and screening and optimization of the selected factors. This systematic review was performed based on the preferred reporting items for systematic reviews and meta-analysis (PRISMA) checklist (14). The present study is a systematic review of in vitro studies because its protocol was not registered.
3. Results
3.1. Summary of the Included Studies and Optimization of Enzyme Production
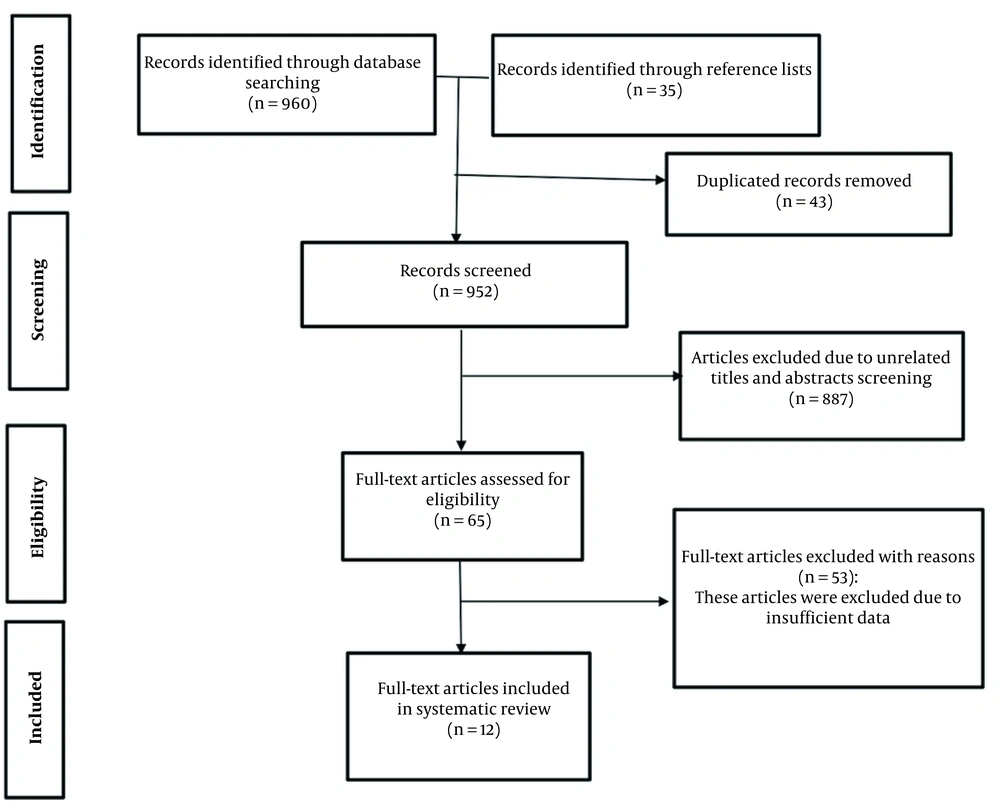
The flow chart of this systematic review for the article selection process is presented in Figure 1. A total of 995 articles were identified in the reference lists of related articles and also our initial search. After 43 duplicate articles were removed, 952 articles were screened based on the title and abstract. Twelve studies were ultimately included in this systematic review, including the optimization of alpha-amylase production in bacteria (seven studies), yeasts (two studies), fungi (two studies), and protists (one study). Box-Behnken design (BBD), central composite design (CCD), and Taguchi methods were the most used statistical designs to calculate process optimization (15-26).
3.1.1. Culture Medium Condition
Several microorganisms, such as bacteria, fungi, yeast, and protist, can produce alpha-amylase; on the other hand, the condition for types of microorganisms is different from optimizing enzyme production. Growth conditions are different for several bacteria types; for instance, the best pH condition for thermophilic Bacillus subtilis is 8.4 (27); however, for Bacillus amyloliquefaciens 04BBA15 is 5.5 (28). Most research on alpha-amylase-producing microorganisms showed that the best temperature for the production of alpha-amylase is around 35°C (17, 18, 27, 29).
Studies on the optimization of alpha-amylase production by fungi indicated that the best condition to optimize alpha-amylase production is the temperature of 25°C, incubation time of 120 hours, and pH of 7.2 (21, 23, 30). The highest alpha-amylase production in yeast was observed in the pH range from 5.5 to 6 and during incubation of 72 hours (24, 31, 32). A summary of the descriptive culture medium condition of included studies is provided in Table 1.
| Author | Microorganism | Statistical Design | Alpha-Amylase Activity | Optimized Growth Condition |
|---|---|---|---|---|
| Du et al. (15) | Bacillus amyloliquefaciens BH1 | Central composite design | 198.26 U/mL | Soluble starch: 12.51 g/L; Na2HPO4: 60.82 g/L; initial pH: 6.81 |
| Gangadharan et al. (16) | Bacillus amyloliquefaciens | Box-Behnken design | 965.9 U/mL | Substrate concentration: 12.5%; incubation period: 42 h; CaCl2: 0.0275 M |
| Panda et al. (17) | Lactobacillus plantarum MTCC 1407 | Central composite design | 4022 U/min/mL | Incubation period: 36 h; pH: 7.0; temperature: 35°C |
| Sharma and Satyanarayana (18) | Bacillus acidicola | Central composite design | 7440 ± 200 IU l - 1 | Starch: 2.75%; K2HPO4: 0.01%; inoculum size: 2% (v/v); temperature: 33°C |
| Uma Maheswar Rao and Satyanarayana (19) | Geobacillus thermoleovorans | Central composite design | 29690 U/L | Glucose: 3%; riboflavin: 200 μg; inoculum density: 3% |
| Uma Maheswar Rao and Satyanarayana (19) | Bacillus subtilis RSKK96 | Taguchi experimental design | 503.26 U/mg | Casein: 1%; corn meal: 1%; glutamic acid: 0.01%; incubation time: 72 h; inoculum density: 1% |
| Gigras et al. (21) | Aspergillus oryzae | Central composite design | 161 U/mL | pH: 7.3; incubation time: 120 h; starch: 6.92 g/L, yeast extract: 4.9 g/L KH2PO4 |
| Kammoun et al. (22) | Aspergillus oryzae CBS 819.72 | Box-Behnken design | Not reported | Temperature, agitation, and inoculum size |
| Naili et al. (23) | Aspergillus oryzae S2 | Box-Behnken design | 750 U/mL | Temperature: 24°C; urea:1 g/L; C/N ratio: 2 |
| Kar and Ray (24) | Streptomyces erumpens MTCC 7317 | Central composite design | 4143 U | Incubation period: 36 h; pH: 6; temperature: 50°C |
| Manivasagan et al. (26) | Streptomyces sp. MBRC-82 | Box-Behnken design | 163.98 U/mL | Soluble starch: 5.8484 g; peptone: 3.5191 g; NaCl: 0.3829 |
| Shirodkar and Muraleedharan (26) | Ulkenia sp. | Central composite design | 71.93 U/mL | Glucose: 0.21%; substrate: 0.21%; yeast extract: 0.33% |
4. Discussion
According to our knowledge, this is the first systematic review to assess the alpha-amylase production and purification from the microbial source by statistical design. Ten different microorganisms had been reported in the selected studies, including B. amyloliquefaciens (15, 16), Lactobacillus plantarum (17), Bacillus acidicola (18), Geobacillus thermoleovorans (19), Bacillus megaterium (33), B. subtilis (20), A. oryzae (21-23), Aspergillus flavus (23), Streptomyces erumpens (24, 25), and Ulkenia sp. (26). Bioprocess factors, including physical conditions and medium composition, are applied for optimized alpha-amylase production. Plackett-Burman design is employed to find the most effective factors for alpha-amylase production. Six studies had used the Plackett-Burman design for screening the significant factors. Significant factors for alpha-amylase production are substrate concentration, incubation period, CaCl2 for the bacteria, temperature, agitation and inoculum size, urea concentration, C/N ratio for fungi, and soluble starch, peptone, NaCl for yeasts.
Several statistical experimental designs, including BBD, CCD, and Taguchi experimental design, have been used in bioprocess optimization of alpha-amylase production from the microorganisms. Seven out of thirty studies had used the central composite design (15, 17-19, 21, 24, 26), Box-Behnken was employed in five studies (16, 22, 23, 25, 33), but only one study optimized alpha-amylase production using Taguchi experimental design (20).
Starch concentration (15, 16, 18, 33) and pH (15, 17, 33) are the most repeated significant factors in the research to optimize the alpha-amylase-concentration of the bacteria. Starch is known as a carbon source, and due to improving the population of alpha-amylase-producing bacteria, this factor can increase enzyme production (34). The optimum pH improves the production and secretion of alpha-amylase. Also, bacteria require a neutral pH (around seven) (35), and the best pH for alpha-amylase production is around seven. In the eukaryotic cell, the most repeated significant factor is temperature. The temperature is the most important factor in the production of alpha-amylase by eukaryotic cells (22-24). Also, the temperature is an important factor that directly affects microorganisms’ growth and subsequently increases alpha-amylase production (36). The best temperature for alpha-amylase production is directly related to microorganism type; however, the best temperature for enzyme production for fungi is 24°C.
4.1. Conclusions
This systematic review discussed alpha-amylase production by several types of microorganisms, including B. amyloliquefaciens, L. plantarum, B. acidicola, G. thermoleovorans, B. megaterium, B. subtilis, A. oryzae, A. flavus, and S. erumpens. The most significant factors for alpha-amylase production by the bacteria were pH and starch concentration, and for the eukaryotic cell (fungi, yeasts, and Ulkenia sp.) was temperature. The starch concentration for alpha-amylase production is variable by several microorganisms, but the best pH for enzyme production is around seven. Also, the best temperature for fungi to produce alpha-amylase is 24°C. The results showed that the growth conditions (such as pH, temperature, and starch concentration) need to be optimized to increase alpha-amylase yield.