1. Background
Staphylococcus aureus is the gram-positive bacterium that causes many infections and syndromes. Staphylococcus aureus is the most common agent of nosocomial infections. The highest rate of infections due to S. aureus were seen among hospitalized patients with predisposing factors (1-3).
Methicillin-resistant S. aureus (MRSA) is commonly emerging in the types of nosocomial and community aquired infections (1-3). The growing up of prevalence of MRSA isolates is an increasing problem (1). In this situation we need to use other antibiotics. Macrolide lincosamide-streptogramin B (MLSB) antibiotics are the therapeutic choices for treatment of infections due to MRSA isolates. Clindamycin also has some advantages, such as less cost and inhibition of production of some toxins and virulence factors in staphylococci (4, 5). Clindamycin is a common antibiotic to treatment of respiratory tract, bone, joint, skin and, soft tissue infections. This antibiotic has low side-effects. Thus, it is appropriate for prolonged therapy (6, 7). Different mechanisms are responsible for bacterial resistance to macrolides including efflux pump, enzymatic antibiotic inactivation and target site modification (5). One of the prevalent mechanism of resistance to clindamycin is inducible resistance (iMLSB) and the most frequent mechanism for inducible resistance in S. aureus is modification in target site by erm (erythromycin ribosome methylase) genes. The treatment by clindamycin in patients with iMLSB resistance phenotype may lead to constitutive MLSB phenotype (cMLSB) and treatment of this infections is failed. The common laboratories cannot detect inducible resistance (erythromycin-resistant and clindamycin-sensitive) by routine laboratory procedure. The common phenotypic method to detect inducible clindamycin resistance is the method is known as D-test because flattening of zone (D-shaped) around clindamycin disk in the area between the two disks (erythromycin and clindamycin) that reveal inducible clindamycin resistance (D-test positive) (7, 8).
2. Objectives
In this study, we evaluated inducible clindamycin resistance in S. aureus strains isolated from hospitalized patients in the Imam Reza Hospital in Kermanshah Province, west of Iran.
3. Methods
3.1. Study Design
This descriptive study was performed on 126 strains of S. aureus strains isolated from hospitalized patients in the Imam Reza Hospital in Kermanshah Province, west of Iran, from June to December 2019.
3.2. Sample Collection and Laboratory Identification
The samples were obtained from all of clinical specimens, such as blood, cerebral spinal fluid (CSF), urine, vaginal, nasal, pus, tracheal and, wound swabs. The samples were inoculated in Blood agar and Mannitol Salt agar plates and incubated at 35°C for 24 - 48 hours. Gram stain, catalase, coagulase and DNAase tests were used for identification of S. aureus isolates. The confirmation of S. aureus isolates was performed by demonstration of femB gene presence using polymerase chain reaction (PCR) method (9).
3.3. Antibiotic Resistance Rate to Clindamycin and Erythromycin
The disk diffusion method was used for determination of the antibiotic resistance rate of isolates against clindamycin and erythromycin based on Clinical and Laboratory Standard Institute (CLSI) guidelines (10). We used clindamycin (2 µg) and erythromycin (15 µg) disks. Antibiotic disks were placed 30 mm apart on surface of the plates. They were incubated at 37°C for 18 - 24 h. Findings on the diameter of the halo created around the antibiotic disk were measured using a millimeter ruler and interpreted based on CLSI guidelines (10).
3.4. Cefoxitin Disk Diffusion Test
For detection of MRSA isolates by phenotypic method, we used cefoxitin disk (30 µg). Inhibition zones diameter of ≤ 21 mm were considered as MRSA (10).
3.5. Inducible Resistance to Clindamycin by D-test
In this test inducible resistance to clindamycin was detected by D-test based on CLSI guidelines (10). Briefly, erythromycin (15 µg) disk was placed apart on 15 mm from clindamycin (2 µg) disk on a Mueller-Hinton agar plate. After overnight incubation at 37°C, flattening of zone (D-shaped) around clindamycin in the area between the two disks, reveal inducible clindamycin resistance (D-test positive). There are three different phenotypes for interpretation of this test.
(1) Inducible MLSB (iMLSB), phenotype (D+): Resistance to erythromycin and sensitive to clindamycin (zone size ≥ 21 mm) with a D-zone of inhibition around the clindamycin disk.
(2) Constitutive MLSB phenotype (cMLSB): Resistance to both erythromycin and clindamycin
(3) MS phenotype: Resistance to erythromycin and sensitive to clindamycin, D-test negative.
(4) The susceptible phenotype (S phenotype): Sensitive to both clindamycin and erythromycin (11).
3.6. PCR Method
All S. aureus isolates were investigated for femB, mecA, ermA, ermB, and ermC genes using specific primers (Table 1) (12-16). Total DNA was extracted using a High Pure PCR Template Preparation Kit (Roche, Basel, Switzerland). PCR amplification was performed using automated thermal cycler (Applied Biosystems, USA). The used PCR conditions were including: Initial denaturation at 95ºC for 5 minutes, denaturation at 1 minute, annealing at different degrees for each gene (Table 1) for 1 minute, extension at 70°C for 1 minute for 35 cycles and final extension at 70°C for 10 minutes. Gel electrophoresis of PCR products carried out in 1.5% agarose gel at 85 V for 45 minutes and visualized on an ultraviolet (UV) transilluminator (BioRad, USA).
| Genes and Primers | Sequences (5' to 3') | Product Size, bp | Annealing Temperature (°C) | References |
|---|---|---|---|---|
| mecA | 147 | 62 | 14 | |
| F | GTGAAGATATACCAAGTGATT | |||
| R | ATGCGCTATAGATTGAAAGGAT | |||
| femB | 388 | 55 | 15 | |
| F | CGTGAGAATGATGGCTTTGA | |||
| R | TTAATACGCCCATCCATCGT | |||
| ermA | 190 | 54 | 12 | |
| F | AAGCGGTAAACCCCTCTGA | |||
| R | TTCGCAAATCCCTTCTCAAC | |||
| ermB | 142 | 55 | 13 | |
| F | CTATCTGATTGTTGAAGAAGGATT | |||
| R | GTTTACTCTTGGTTTAGGATGAAA | |||
| ermC | 297 | 52 | 16 | |
| F | AATCGTCAATTCCTGCATGT | |||
| R | TAATCGTGGAATACGGGTTTG |
Abbreviations: F, forward primer; R, reverse primer.
3.7. Data Analysis
Statistical calculations were performed using SPSS software version 16 for descriptive statistics of data. Statistical significance of differences between findings was evaluated by chi-square (χ2) test. A P-value < 0.05 was considered statistically significant.
4. Results
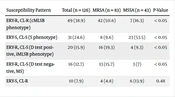
In this study 126 isolates were confirmed as S. aureus using phenotypic methods and presence of femB gene. 53 cases (42.6%) and 73 cases (57.4%) were isolated from males and females, respectively. According to presence of mecA gene, 83 cases (65.9%) were MRSA and 43 cases (34.1%) were methicillin sensitive S. aureus (MSSA). The resistance rate against erythromycin and clindamycin were 67.4% and 52.2%, respectively. In all S. aureus isolates 49 cases (38.9%) of isolates resistant to both erythromycin and clindamycin indicating constitutive MLSB phenotype (cMLSB phenotype); 31 cases (24.6%) isolates were sensitive to both erythromycin and clindamycin, 20 cases (15.9%) isolates showed positive D-test indicating inducible MLSB phenotype (iMLSB phenotype), while 16 cases (12.7%) were negative for D test indicating MS phenotype, 10 cases (7.9%) isolates were sensitive to erythromycin and resistance to clindamycin (S phenotype). The rate of inducible clindamycin resistance in MRSA isolates was higher than in MSSA isolates (P-value < 0.05). The rate of cMLSB phenotype and iMLSB phenotype among MRSA isolates were 50.6% and 19.3%, respectively. Among MSSA isolates rate of cMLSB phenotype and iMLSB phenotype were 16.3% and 9.3%, respectively (Table 2).
| Susceptibility Pattern | Total (n = 126) | MRSA (n = 83) | MSSA (n = 43) | P-Value |
|---|---|---|---|---|
| ERY-R, CL-R (cMLSB phenotype) | 49 (38.9) | 42 (50.6) | 7 (16.3) | < 0.05 |
| ERY-S, CL-S (S phenotype) | 31 (24.6) | 8 (9.6) | 23 (53.5) | < 0.05 |
| ERY-R, CL-S (D test positive, iMLSB phenotype) | 20 (15.9) | 16 (19.3) | 4 (9.3) | < 0.05 |
| ERY-R, CL-S (D test negative, MS) | 16 (12.7) | 13 (15.7) | 3 (7) | < 0.05 |
| ERY-S, CL-R | 10 (7.9) | 4 (4.8) | 6 (13.9) | 0.48 |
Abbreviations: ERY, erythromycin; CL, clindamycin; S, sensitive; R, resistant; MRSA, methicillin resistant S. aureus; MSSA, methicillin susceptible S. aureus; S phenotype, sensitive to both erythromycin and clindamycin; cMLSB, constitutive resistance macrolide, lincosamide, and streptogramin B; iMLSB, inducible resistance macrolide, lincosamide, and streptogramin B; D test positive, MS phenotype: resistant to erythromycin and sensitive to clindamycin; D test negative, S phenotype, sensitive to both erythromycin and clindamycin.
a Values are expressed as No. (%).
These iMLSB resistance phenotype isolates were investigated for the presence of the erm genes. The ermC and ermA genes were showed in 7 cases (35%) and 4 cases (20%) isolates, respectively. All 11 positive cases for erm genes, were isolated from MRSA isolates. The ermB gene is not detected in any cases and, 9 cases (45%) isolates did not have any erm genes.
5. Discussion
Staphylococcus aureus is the most common agent of nosocomial infections. In recent years, prevalence of MRSA isolates has been increased and treatment of infections due to these isolates has been harder than before (1). In this situation we need to use other antibiotics. Macrolide Lincosamide-Streptogramin B antibiotics are the therapeutic choices for treatment of infections due to MRSA isolates. Clindamycin also has some advantages such as less cost and inhibition of production of some toxins and virulence factors in staphylococci (4, 5). The presence of clindamycin resistance phenotype in S. aureus clinical isolates could render effectiveness of this antibiotic for treatment of infections due to S. aureus isolates. This resistance may be constitutive or inducible (7, 8). In this present study we aimed to evaluate prevalence of resistance pattern to erythromycin and clindamycin and also erm genes occurrence in S. aureus isolates.
In this study, we used a D-test for phenotypically detection of several susceptibility pattern using erythromycin and clindamycin disks that located apart on surface of culture medium. There are several different phenotypes for interpretation of this test. Totally cMLSB, iMLSB, S and MS phenotypes were seen in 38.9%, 15.9%, 24.6 and 12.7% of S. aureus isolates, respectively.
In this study, iMLSB phenotype D-test positive (resistance to erythromycin and sensitive to clindamycin) was seen in 15.9% isolates. This result is higher than the result of other studies were conducted by Kilany in Egypt and Rahbar and Hajia in Iran that iMLSB phenotype were detected in 7.7% and 10.8% of S. aureus isolates, respectively (17, 18), and lower than other previous studies were performed by Raut et al. and Bobenchick et al. that they reported iMLSB phenotype in 25.6% and 22.3% isolates, respectively (19, 20). In some studies, the iMLSB rate was reported very high (82% and 88%) (21, 22). Findings of this study revealed the prevalence of the iMLSB phenotype between MRSA and MSSA isolates was statistically different (P < 0.05). Resistant strains with iMLSB phenotype can lead to cMLSB phenotype and cause failure in treatment with clindamycin. The most prevalent clindamycin resistant phenotype in this study was cMLSB phenotype (resistance to both erythromycin and clindamycin) (38.9% of isolates) and this finding showed that cMLSB phenotype was higher than iMLSB phenotype similar to other study was conducted by Mahesh et al. (23). In the present study, the prevalence of cMLSB phenotype in MRSA isolates was significantly more than MSSA isolates, which in consistent with studies were performed in other countries (23, 24). The high frequency of cMLSB phenotype in MRSA isolates may be due to the selection pressure after therapeutic failure of methicillin and utilization of erythromycin and clindamycin. The iMLSB strains may be changed to cMLSB, thus laboratories have to detect iMLSB strains by D-test and eradicate these strains by effective therapeutic agents. In present study, MS phenotype (resistance to erythromycin and sensitive to clindamycin, D-test negative) was observed in 12.7% S. aureus isolates. In accordance to our study, the prevalence of MS phenotype is low in other study (25).
We investigated frequency of erm genes (ermA, ermB and ermC) in S. aureus isolates with iMLSB phenotype by PCR method. Our findings of this study showed of 20 MRSA isolates with iMLSB phenotype, the ermA and ermC genes were found in 4 cases (20%) and 7 cases (35%) isolates, respectively. The ermB gene is not detected in any cases. The ermC gene was the most prevalent gene in S. aureus isolates with iMLSB phenotype in present study. This result is similar to many studies revealed that ermC gene was associated with the majority of resistance to erythromycin among the MRSA isolates (26, 27). Contrary to results of this study, in some studies, ermA gene more frequent than ermC gene such as the study were conducted by Saderi in Iran that he reported prevalence of ermA and ermC in 60.3% and 54.8% of isolates that these results are higher than our findings in this study (28). Schmitz detected the ermA gene in 67% isolates and the ermC gene in 23% (29). In Korea, Jung et al. identified ermA gene in 89% and ermC gene in 5% isolates (30). In Iran, Moosavian et al. detected the ermA and ermC genes in 41.1% and 17.7% of S. aureus isolates, respectively (31). The ermB gene is not detected in any cases in this study. Cetin et al similar to our study found no ermB gene in S. aureus isolates (32). Other study reported low prevalence of ermB gene (33). The ermB gene usually detected in staphylococci spp. of animal origin and spread between streptococci and enterococci (33). These difference in prevalence of S. aureus isolates with iMLSB phenotype may be correlated to patients age, geographical area, rate of using antibiotics, bacterial species, source of specimens and community or nosocomial infections. The presence of other mechanisms of resistance leading to the complexity of resistance in S. aureus to MLSB antibiotics.
5.1. Conclusions
In general, results of this research showed high frequency of resistance to clindamycin and erythromycin among S. aureus isolates and cMLSB to be the most pattern phenotype. The ermC gene is the most isolated gene. Because variation of antimicrobial resistance pattern in geographic regions, obtaining local results is useful for detecting and more appropriate control of nosocomial infection due to S. aureus isolates. We recommended conducting the D-test for detection of iMLSB phenotypes in microbiology laboratories because the D-test is simple and cheap method that can be done in every routine laboratory.