1. Background
Cancer remains a significant global health challenge, contributing to nearly 10 million deaths in 2020 (1). Despite advancements in cancer diagnosis and treatment, numerous obstacles persist, including drug resistance, adverse effects, and high treatment costs (1). Hence, there is an imperative to explore new reservoirs of anticancer agents that are efficacious, safe, and economically viable. Marine organisms, particularly sponges, represent a vast reservoir of bioactive compounds with potential anticancer properties (2, 3). These compounds, sourced from marine plants, algae, bacteria, actinomycetes, fungi, and soft corals, have demonstrated promising outcomes in both in vitro and in vivo studies (4). Several marine-derived molecules are presently undergoing various stages of clinical trials for anticancer therapy, highlighting the promise of these natural products in cancer treatment (5).
E3 ubiquitin ligases, such as Mdm2, play a pivotal role in cancer initiation and progression (6, 7). They contribute to critical cancer characteristics like sustained proliferation, immune evasion, and apoptosis (7). Particularly, Mdm2 represents a potential drug target and prognostic biomarker in melanoma (8). The interaction between Mdm2 and P53, a tumor suppressor, is a central focus in cancer research, and natural products have displayed potential in modulating this interaction (9).
Poly(ADP-ribose) polymerase 1 (PARP-1), pivotal in DNA repair and genomic stability, is often overexpressed in various cancers, rendering it a plausible therapeutic target (10). Inhibiting PARP1 can sensitize cancer cells to DNA damage and bolster the effectiveness of radio- and chemotherapy (10). This is particularly pertinent concerning P53, as PARP1 inhibitors have the potential to amplify the apoptotic response mediated by p53 (11). Moreover, the synthetic lethality of PARP1 inhibitors in BRCA1/2-deficient model systems underscores their promise in cancer treatment (12). However, PARP1's role in cancer is multifaceted, with evidence suggesting both oncogenic and tumor-suppressive functions (13).
Hence, natural products targeting Mdm2, PARP1, and P53 hold considerable promise in cancer therapy.
Marine sponges collected from the Persian Gulf have demonstrated antioxidant properties, with the methanolic extract of Pseudosaberites clavatus exhibiting the highest scavenging activity (10). These sponges also harbor antiangiogenic compounds with potential implications in cancer therapeutics (14). Moreover, marine organisms from the Gulf of Oman, encompassing the Persian Gulf, exhibit robust anti-cancer activity, with certain compounds inducing cell death in breast adenocarcinoma models (15). Specific compounds sourced from marine sponges, such as latrunculins and hydantoins, exhibit promise in inhibiting the growth and invasion of prostate cancer cells (16). These findings collectively suggest that marine sponges from the Persian Gulf may indeed possess anti-cancer properties.
Dysidea avara, a marine sponge, holds promise in cancer treatment owing to its bioactive compounds. Sponges of the genus Dysidea belong to the class Demospongiae, order Dictyoceratida, and family Dysideidae (17). Baguley and Wilson (18) and McKeage (19) both discuss the potential of DMXAA, a compound derived from the sponge, in cancer treatment. DMXAA has shown productive interactions with radiation, hyperthermia, and chemotherapeutic drugs, demonstrating efficacy in combination with docetaxel in advanced prostate cancer. Turrini (20) further supports the potential of natural products in cancer treatment, highlighting the anticancer effects of polyphenols present in D. avara. Previous studies have indicated that sponges within the Dysidea spp. possess secondary metabolites with diverse biological properties (21-26). These studies collectively suggest that D. avara, along with its bioactive compounds, holds promise in the development of new anticancer strategies.
Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) is a compound with potential biological activities. It has been suggested to possess antioxidant and cytoprotective capabilities, although these have not been fully validated in vivo (27). Other related compounds, such as ergosta-4,6,8(14),22-tetraen-3-one, have been reported to exhibit diuretic, cytotoxic, antitumor, and immunosuppressive activities (28). Additionally, the compound's potential role in mitochondrial biogenesis, oxidative phosphorylation, and metabolic diseases, particularly diabetes, has been proposed (29). Further research is warranted to comprehensively elucidate the biological activities of ergosta-14,22-dien-3-ol.
2. Objectives
This research aims to investigate the cytotoxic and molecular effects of ergosta-14,22-dien-3-ol (3β, 5α, and 22E) fraction, a natural compound isolated from D. avara, on cancer cells. The specific objectives include evaluating the compound's cytotoxicity against various cancer and normal cell lines, exploring its mechanism of action by measuring the expression of the P53 protein, and assessing its binding affinity and interaction mode with its target protein using computational methods. This study will reveal the anti-cancer potential of the ergosta-14,22-dien-3-ol (3β,5α, and 22E) fraction and will support the development of novel natural-based cancer therapeutics.
3. Methods
3.1. Sampling and Identification
Dysidea avara (Figure 1) was manually obtained from depths of 25-30 meters in the coastal waters of Hengam Island in the Persian Gulf, geographically located at N55°54′55′′-E 55°54′40′′ and N26°41′15′′-E 26°36′43′′. Marine sponges were rinsed using freshwater and distilled water to remove microscopic organisms. Samples were frozen at -20°C before being transferred to a laboratory. Taxonomic identification was achieved using the Hooper identification key, dissociated spicule mounts, light optical microscopes, scanning electron microscopes, and skeleton slides. The scientific name of the sponge sample was confirmed by the Persian Gulf and Oman Sea Ecological Research Organization (30).
3.2. Extraction and Isolation
The D. avara samples underwent initial extraction using a 1:1 mixture of methanol and dichloromethane for 48 hours, yielding concentrated extracts. The residual aqueous phase was then adjusted to 200 mL and partitioned against a 3:2 mixture of dichloromethane and ethanol (31). This process resulted in the formation of organic layers, which were evaporated to produce the sub-extracts SEA, SEB, SEC, SEE, and SEF with respective weights of 9.04 g, 27.12 g, 1.34 g, 0.414 g, and 0.34 g.
These sub-extracts were screened for steroids using thin-layer chromatography (TLC) with a vanillin reagent. The fraction that showed positive results, SEB, was further separated using column chromatography on a silica gel column with a gradient of n-hexane and ethyl acetate from 100:0 to 0:100. This yielded 15 fractions (SEB-1 to SEB-15), which were also tested for steroids.
Fraction SEB-11 (1.45 g) underwent further purification via silica gel column chromatography. A gradient elution system was employed, using a mixture of ethyl acetate and methanol with increasing polarity (from 100:0 to 0:100). The subfraction SEB11-9, eluted with a 90:10 ethyl acetate-methanol solvent system, exhibited a band with a positive response to the Vanillin-sulphuric acid test for steroids. This band was isolated using thin-layer chromatography (TLC) (32). Following isolation by TLC, the band was dissolved in acetone and filtered, yielding the purified compound. The structure of the isolated compound was then elucidated using Gas Chromatography-Mass Spectrometry (GC/MS) analysis. The analysis was performed on an Agilent 7000 Series Triple Quad GC/MS Mainframe model with helium as the carrier gas and a C5975 detector, located at the Shahid Beheshti University research laboratory.
3.3. Cytotoxicity Assay
The T-lymphocytic leukemia cell line (Jurkat/E6-1) and the normal human embryonic kidney cell line (Hek293) were provided by the Pasteur Institute of Iran, and both cell lines were cultured in vitro as monolayers. The RPMI-1640 media (Gibco) were supplemented with 0.6 mg/mL glutamine, 200 IU/mL penicillin, 200 IU/mL streptomycin, and 0.1 mg/mL gentamycin. For the Sodium 3-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene sulfonic acid hydrate (XTT) assay, cells were trypsinized and centrifuged at 1500 rpm for 5 minutes before being distributed into 96-well plates at a density of 5×103 cells per well. A volume of 200 μL of RPMI-1640 medium was added to each well, and the plates were then incubated for 24 hours in a CO2 incubator while the cell density was monitored using inverted microscopy. Following 48 hours of incubation to ensure cell attachment to the plate, the medium was replaced with 100 μL of fresh medium containing varying concentrations (2, 10, 50, 100, and 200 μg/mL) of the Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) fraction. After 24 hours, the results were assessed, and the experiment was conducted in triplicate. The samples were then incubated with 50 μL of XTT solution (1 mg/mL) for 6 hours, and optical density (OD) was measured at wavelengths of 490 nm and 690 nm using the Bio-Tek ELx 800 ELISA Reader model. The IC50 value was determined using a graph of percentage cell viability, representing the concentration at which 50% of the cells are killed (33).
3.4. Western Blot Analysis
The Jurkat/E6-1 and Hek293 cells were grown in 6-well plates and treated with different concentrations of Ergosta-14, 22-dien-3-ol (3β, 5α, 22E) after 24 hours of incubation at 37 °C and 5% CO2. The protein concentration was measured using a bicinchoninic acid (BCA) protein quantitation kit and subjected to SDS-PAGE (12% polyacrylamide), followed by transfer to PVDF membranes. After blocking the membranes with 5% skimmed milk-containing TBS-T solution (Tris-buffered saline with 0.1% Tween 20), the membranes were incubated with primary anti-Tumor protein P53 (P53) and anti-β-actin antibodies overnight at 4°C (34). The membranes were then incubated with the horseradish peroxidase (HRP)-labeled anti-rabbit immunoglobulin G (IgG) and visualized using a Chemi-Doc gel documentation system with chemiluminescence (ECL) kit. The blots were quantified using ImageJ software.
3.5. Molecular Docking Studies
The 2D structure of Ergosta-14,22-dien-3-ol (3β,5α, and 22E) was drawn using ChemBioDraw 12.0 and then minimized with MM+ and AM1 methods in Hyperchem 8 (35). Next, automated docking was performed with AutoDock 4.2.5.1 and ADT 1.5.6 using the DOCKFACE script (36). Subsequently, the crystal structures of Poly (ADP-ribose) polymerase-1 (PARP1) (3L3M) (37) and E3 ubiquitin-protein ligase (MDM2) (1T4E) (38) were downloaded from the Protein Data Bank. The proteins were prepared with ADT, and the grid maps were generated with AutoGrid (36). The grid sizes for 1T4E and 3L3M were 40 × 40 × 40 and 74 × 80 × 82, respectively, with 0.375 Å spacing. The grid centers for 1T4E and 3L3M were (43.807, 12.419, and 28.821) and (24.875, 11.115, and 26.683), respectively. The docked results were then clustered with a 2 Å RMSD cutoff. For internal validation, the co-crystal ligands of PARP1 (3L3M) and MDM2 (1T4E) were used as references. Moreover, 100 docking runs were executed with the Lamarckian genetic algorithm, and the protein-ligand interactions were analyzed with Chimera 1.13 (39) and LigPlot+ (40). Finally, the docking results were evaluated based on energy and interactions between Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) and the targets.
3.6. Pharmacological Properties of Absorption, Distribution, Metabolism, Excretion, and Toxicity
The pharmacokinetic profile of Ergosta-14,22-dien-3-ol (3β, 5α, 22E), including absorption, distribution, metabolism, excretion, and toxicity (ADMET), was predicted using pkCSM (41) and SwissADME (42).
3.7. Statistical Analysis
The mean and standard deviation of three independent experiments were used to express the results. Statistical analysis was performed on the IC50 values obtained from the XTT assay. The Kolmogorov-Smirnov and Shapiro-Wilk tests were used to determine whether the IC50 values followed a normal distribution. The differences between groups were assessed by one-way analysis of variance (ANOVA) followed by Tukey's post hoc test for multiple comparisons. The statistical analysis was conducted using GraphPad Prism software (version 9).
4. Results
4.1. Isolation and Verification of Steroids
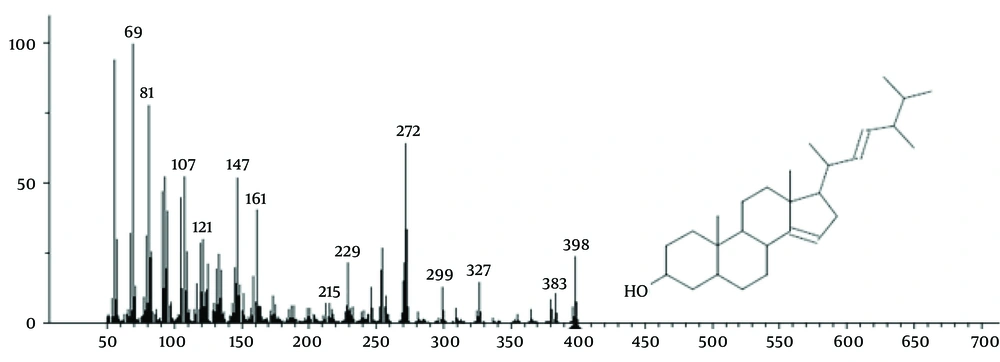
The isolated fraction was confirmed to be Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) with the chemical formula C28H46O (structure shown in Figure 2). This steroid compound exhibited a molecular weight of 428.64 g/mol and was detected at 90% purity in the SEB11-9 fraction (eluted with a 90:10 ethyl acetate-methanol solvent system) at retention times between 40 and 48 minutes (Figure 2).
4.2. Cytotoxicity Assay
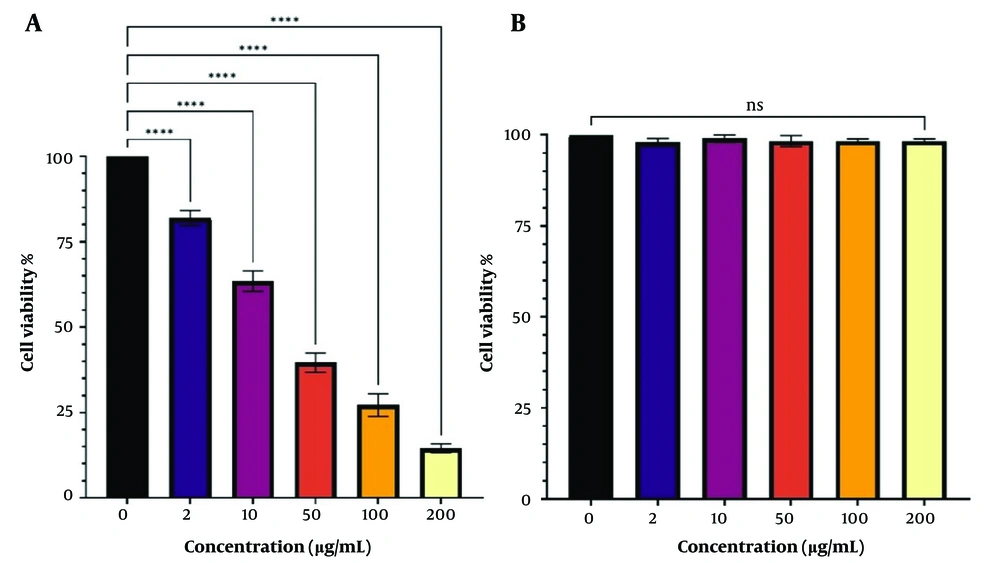
The XTT assay was used to evaluate the cytotoxicity of Ergosta-14,22-dien-3-ol (3β,5α, and 22E) on Jurkat/E6-1 and Hek293 cells. The response of Jurkat/E6-1 cells to increasing concentrations of Ergosta-14,22-dien-3-ol (3β,5α, and 22E) was exponential, with a significant decrease in viability observed at concentrations of Ergosta-14,22-dien-3-ol (3β,5α, and 22E), and a further decline at the highest concentrations tested. The estimated IC50 value for Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) was 26.59 μg/mL for Jurkat/E6-1 cells. The in vitro dosage- and time-dependent activity of Ergosta-14,22-dien-3-ol (3β,5α, and 22E) against cultured human embryonic kidney (Hek293) cells is shown in Figure 3B, and cytotoxic activity was not observed.
A, Cytotoxic assay of Ergosta-14, 22-don-3-ol, (3β.5α, 22E) treatment in Jurkat/ E6-1; and B, Hek293. At 24 h of treatment, the activities of Ergosta-14, 22-don-3-ol, (3β.5α, 22E) against the viability of treated cells were evaluated through the XTT assay. The results are presented as mean ± SEM for triplicates. (ns not significant and **** P < 0.0001 vs untreated control).
4.3. Western Blot Analysis
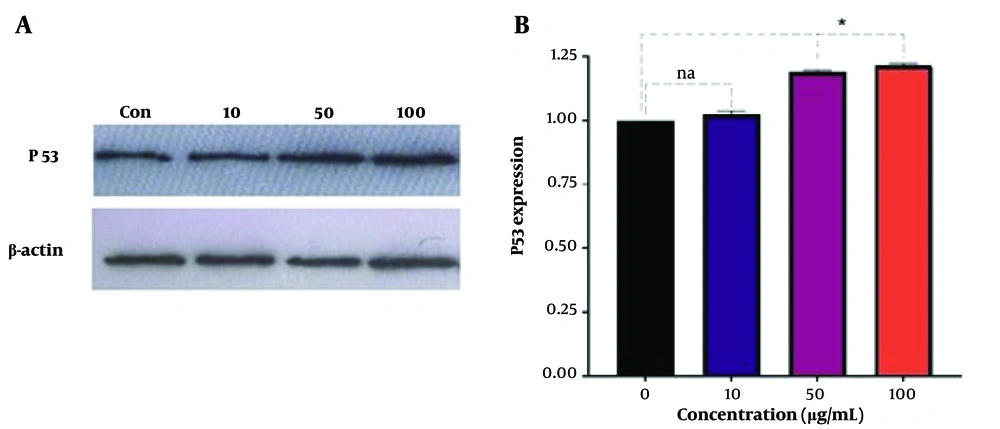
The study investigated the activity of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) treatment on the expression of P53 in Jurkat/E6-1 cell lines. The cells were treated with 10, 50, and 100 µg/mL of Ergosta-14,22-dien-3-ol for 24 hours, and the expression of P53 and β-actin proteins was analyzed using Western blotting. The results, shown in Figure 4, indicate a significant increase in P53 expression in cells treated with 50 and 100 µg/mL of Ergosta-14,22-dien-3-ol compared to the untreated control group.
Cells were treated with concentrations of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) (10, 50, and 100 µg/mL) for 24 hours (controls remained untreated). In this study, β-actin served as an internal control. Pixel density values are displayed for P53. ImageJ software was used to determine the quantitative levels of P53 after Western blot analysis. The results are presented as mean ± SEM for triplicates. (Na not significant and *P < 0.05 vs. untreated control). P53 gene expression.
4.4. Molecular Docking Activities
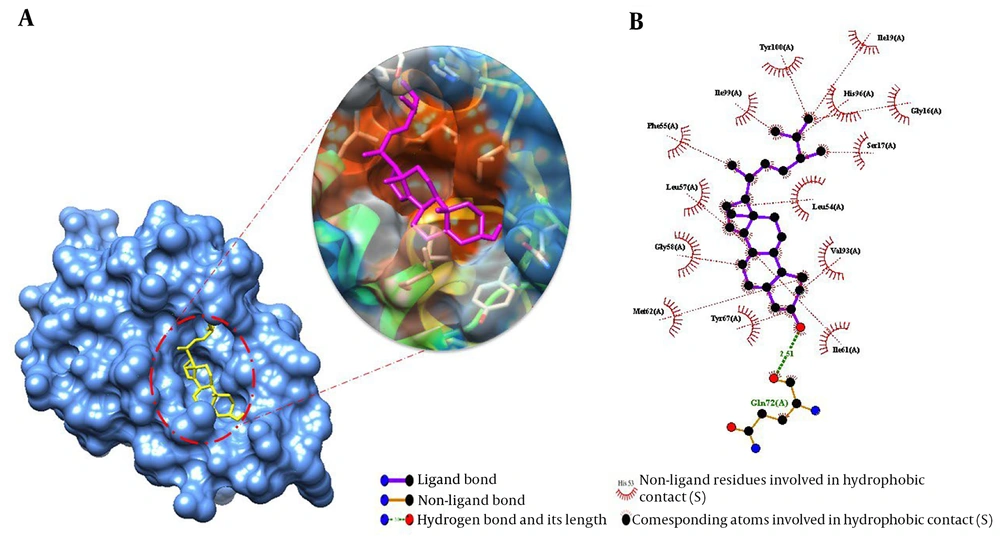
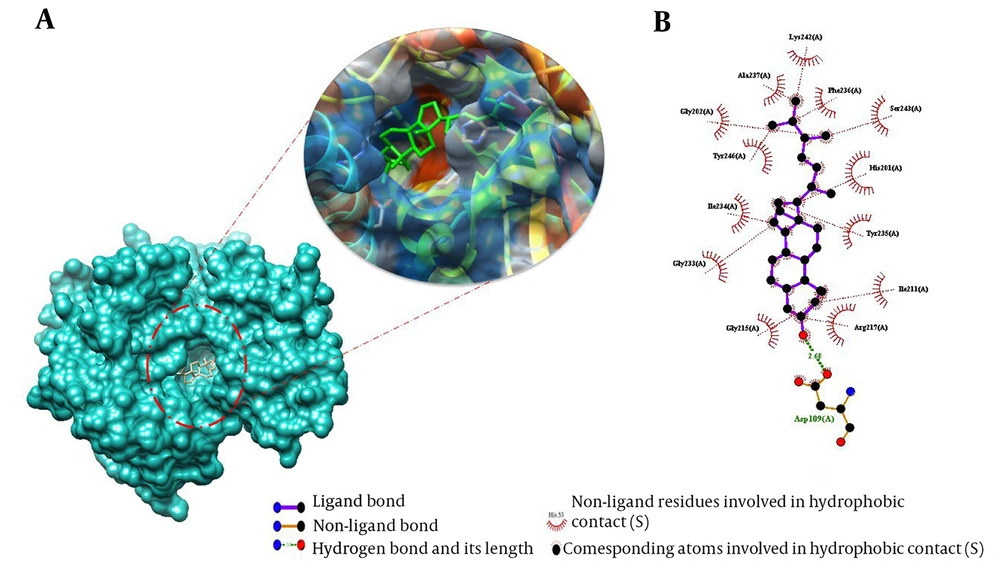
This study used molecular docking simulations to explore the binding mode of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) with PARP and MDM2. The free binding energy (ΔG bind) and the critical residues involved in the interactions were calculated and reported in Table 1 and Figures 5 and 6. The aim was to understand the molecular mechanism of action of this compound. Table 1 shows that the best-docked poses had ΔG bind values ranging from -12.78 kcal/mol for 3l3 m to -10.50 kcal/mol for 1t4e. The compound had higher docking affinity than the co-crystal ligands, except for 1t4e. These results suggest that Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) had the strongest binding, especially for 3l3m. Appendix 1 shows the compound formed a hydrogen bond with Gln72A and van der Waals contacts with several other residues. The docking simulations of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) with 3L3M showed a hydrogen bond with Asp109A and hydrophobic interactions with His201A and Tyr246A. The docking procedure was validated by redocking the natural PARP1 (3L3M) and MDM2 (1t4e) substrates. The RMSD values of the redocked ligands were 0.08 and 0.06, respectively. Appendices 1 and 2 show the hydrogen bonds and van der Waals contacts of the redocked ligands with the active site residues. These findings confirm the reliability of the docking method and the binding mode of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) with its targets.
Molecular docking studies of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) against MDM2 (1T4E). Binding mode and molecular interaction of hit ligands with 1T4E. A, The surface representation of 3L3M shows the binding mode of the docked compound. B, Ligplot + profile of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) docking on PDB structures of MDM2. The figure illustrates the 3D and 2D binding modes of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) with the key amino acid residues of 1T4E.
Molecular docking studies of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) against poly (ADP-ribose) polymerase-1 (PARP1) (3L3M). Binding mode and molecular interaction of hit ligands with 3L3M. A, The surface representation of 3L3M shows the binding mode of the docked compound;. B, ligplot + profile of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) docking on PDB structures of PARP1. The figure illustrates the 3D and 2D binding modes of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) with the key amino acid residues of 3L3M.
| Receptors | Parameters | ||||
|---|---|---|---|---|---|
| Binding Energy, kcal/mol | Number of Hydrogen Bonds | Amino Acid Residues | Number of Hydrophobic Interactions | Amino Acid Residues | |
| 3l3m | -12.78 | 1 | Asp109A | 13 | Arg217(A), Ile211(A), Tyr235(A), His201(A), Ser243(A), Phe236(A), Lys242(A), Ala237(A), Gly202(A), Tyr246(A), Ile234(A), Gly233(A), Gly215(A) |
| 1t4e | -10.50 | 1 | Gln72A | 14 | Val93(A), Ile61(A), Ser17(A), Gly16(A), Leu54(A), His96(A), Ile19(A), Tyr100(A), Ile99(A), Phe55(A), Leu57(A), Gly58(A), Met62(A), Tyr67(A). |
4.5. Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis
The pharmacokinetic profile of a compound specifies its properties of Absorption, Distribution, Metabolism, and Excretion (ADME), whereas toxicity defines a compound's safety profile. Small structural changes can have a significant impact on drug candidates' pharmacokinetic and toxicity properties. Experimental analysis of small molecules' pharmacokinetics and toxicity properties is both time-consuming and expensive and does not always scale accurately between animal and human models. The SwissADME tool is used to assess Drug-Likeness and molecule properties, while the pkCSM tool comprehensively characterizes the pharmacokinetic and toxicity properties of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E). The molecule properties, drug-likeness, and ADMET details of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) are shown in Appendix 3.
5. Discussion
The emergence of drug-resistant isolates has increased the need for novel natural compounds. The results of the present study indicate that the fraction containing the compound Ergosta-14,22-dien-3-ol (3β,5α,22E) extracted from D. avara species sponge has cytotoxic effects on lymphocyte cell lines at concentrations of 26.59 µg/mL. However, it does not exhibit lethal effects on human embryonic kidney cells, indicating that the fraction containing the compound Ergosta-14,22-dien-3-ol (3β,5α,22E) selectively targets cancer cells. An experiment was performed on compounds (E)-tetracos8-en-5-ynoic acid (2) and (22E)-ergosta-5,8,22-trien-7-one-3b-ol extracted from the marine Biemna ehrenbergi from the Red Sea using the MTT assay. The findings showed that both compounds had a significant lethal effect on colon cancer cells (HCT-16) at 40 and 45 μg/mL (43). Karthikeyan et al. (44) conducted a study to examine the impact of ethyl acetate ester extract from Saccostrea glomerata containing Ergosta-5,22-dien-3-ol (3β,22E) on breast cancer epithelial cells. The findings revealed that the extract had a lethal effect on cancer cells at a concentration of 100 µg/mL.
Another study indicated that isolated steroid compounds of Dysidea sp. demonstrated cytotoxic activities against human epidermoid carcinoma cells. Additionally, avarone, isolated from the Chinese marine sponge D. avara, inhibited the activity of four human cancer cell lines: Cervix (HeLa), lung (A549), breast (MDA231), and hepatoma (QGY7703) (45). Khaledi et al. (25) showed that extracts from Persian Gulf D. avara possessed anti-inflammatory activity following seven days of treatment. Mice treated with the extract displayed a dose-dependent and significant improvement compared to controls.
The present study investigated the potential activity of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) on the expression of the P53 protein in Jurkat/E6-1 cell lines. Western blotting analysis showed that cells treated with 50 and 100 µg/mL of the compound significantly increased P53 expression compared to the control. These results suggest that Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) may have a potential role in upregulating P53 expression in cancer cells, which could be a promising strategy for cancer treatment since P53 is a well-known tumor suppressor gene. Its activation can lead to cell cycle arrest and apoptosis in cancer cells (47). Therefore, Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) may have the ability to induce P53 expression, which could potentially be a mechanism for its anti-cancer activity. However, further investigations are required to elucidate and comprehend the mechanism of action of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) on P53 expression and to determine its potential as a therapeutic agent for cancer treatment (46). Additionally, it would be interesting to examine the activity of Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) on other molecular targets associated with cancer development and progression.
Poly (ADP-ribose) polymerase inhibition can potentially be used in cancer treatment through at least two mechanisms: enhancing tumor sensitivity to DNA-damaging chemotherapeutic drugs and producing "synthetic lethality" in PARP-dependent cells (47). Small molecule MDM2 inhibitors can block the interaction of MDM2 and P53 by binding to the region I of the MDM2 protein, resulting in an increase in P53 levels and activation of the P53 signaling pathway (48). As a result, small-molecule MDM2 inhibitors inhibit cell proliferation and increase apoptosis (49, 50). The physicochemical characteristic and pharmacological properties study revealed that Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) could be a potent PARP1 and MDM2 inhibitor. The molecular docking results show that Ergosta-14,22-dien-3-ol (3β, 5α, and 22E)has binding affinities with the active sites of PARP-1 and MDM2. The binding energies were lower than co-crystal ligands, indicating higher affinity, especially for PARP-1 inhibition. These findings are consistent with previous studies, for example, Polybromobiphenyl ether derivatives and monocyclofarnesol-type sesquiterpenes from the Indonesian marine sponge Lamellodysidea cf. herbacea have been identified as PTP1B inhibitors (51). Similarly, a polybromodiphenyl ether from the same sponge has been found to inhibit PTP1B activity (52). These findings suggest that marine sponge compounds may have PARP-1 inhibitory properties.
On the other hand, MDM2 suppresses the tumor-suppressing functions of P53 by promoting its rapid degradation (53, 54). These findings highlight the intricate interplay between PARP-1, MDM2, and P53 in the context of DNA damage repair and tumor suppression. The current results suggest that Ergosta-14,22-dien-3-ol may act through similar mechanisms involving PARP-1 and MDM2 inhibition. The ADME predictions indicate favorable pharmacokinetics for Ergosta-14,22-dien-3-ol. The high gastrointestinal absorption and membrane permeability suggest good bioavailability upon oral administration.
In the present study, Ergosta-14,22-dien-3-ol (3β, 5α, and 22E) was docked onto PARP1 and MDM2 proteins, and the docking score was compared with reference compounds 2-{2-fluoro-4-[(2S)-piperidin-2-yl] phenyl}-1H-benzimidazole-7-carboxamide and DI-Chloro-benzodiazepine. It was found that the free binding energy between the compound and PARP1 (target protein) yielded the best outcome of all the dockings in the study. This study represents the first report of PARP1 and MDM2 docking for Ergosta-14,22-dien-3-ol (3β, 5α, and 22E). These findings provide preliminary evidence for the anti-cancer potential of Ergosta-14,22-dien-3-ol against T-lymphocytic leukemia.
5.1. Conclusions
In the current study, Ergosta-14,22-dien-3-ol (3β, 5α, and 22E), a compound extracted from D. avara, specifically induced the death of Jurkat carcinoma cells at very low concentrations while showing no effect on healthy HEK-293 cells. The results of molecular docking and ADME predictions suggest that further experimental validation is required to confirm the effectiveness, safety, and pharmacokinetics of this steroid derived from a marine sponge. Elucidating its pharmacological mechanisms could facilitate its development as a promising marine anti-cancer agent. Therefore, reproducing D. avara marine sponge and then isolating and purifying these compounds could pave the way for developing potent medicinal and drug supplements for improving cancer patients.