1. Background
Diabetes mellitus (DM) encompasses a group of chronic metabolic disorders characterized by sustained elevations in blood sugar (glucose) levels (1). Hyperglycemia, a hallmark of diabetes, leads to severe complications such as retinopathy, heart disease, kidney failure, and mucormycosis infections. In 2017, 425 million people had diabetes, resulting in 4 million deaths. These numbers are projected to rise, burdening healthcare systems (2-4).
There are three main classifications of DM: Type 1 diabetes (T1DM), type 2 diabetes (T2DM), and gestational diabetes (GDM) (5). Type 2 diabetes, the most prevalent form, is characterized by progressive insulin resistance and declining insulin secretory capacity. Gestational diabetes is a temporary condition during pregnancy that resolves after childbirth (1, 6). Effective diabetes management across all classifications depends on timely diagnosis (7).
Artificial intelligence (AI) is a rapidly evolving field with expanding applications in prediction, risk assessment, and early diagnosis of diabetes. Machine learning (ML) algorithms hold immense potential to revolutionize clinical practice by automating diagnoses (8). Diabetes care is at the forefront of adapting and integrating ML technology, offering promising potential for improving patient outcomes (8, 9).
1.1. Artificial Intelligence in Medicine
This paper delves into the use of AI in medicine. Artificial intelligence, a discipline within computer science, develops systems and methods for data analysis across diverse applications (10, 11). This section provides a brief review of several popular computational intelligence paradigms.
Machine learning is a core AI technique for pattern recognition within specific datasets. Through data fitting, machines can "learn" and apply this knowledge to similar future scenarios (12).
1.2. Applications of Machine Learning in Medicine
Diagnosis of diabetes greatly benefits from ML techniques in medicine. Machine learning aids in identifying high-risk individuals for specific diseases. Recent successes include utilizing ML algorithms to predict diabetes at an early stage using electronic health record data (13).
Machine learning encompasses two prominent forms: Artificial neural networks (ANNs) and deep learning (DL), with the latter being more complex. Inspired by the biological brain's structure and function, ANNs consist of interconnected nodes (neurons) that process information using mathematical functions (14).
Deep learning employs ANNs to model and analyze data. It uses layered nonlinear units to learn intricate patterns from large datasets, eliminating the need for manual feature engineering. Subcategories include convolutional neural networks (CNNs), recurrent neural networks (RNNs), stacked autoencoders (SAEs), and deep belief networks (DBNs) (15). Deep learning approaches like CNNs have been successful in tasks such as predicting diabetic retinopathy from retinal scans, highlighting their potential to improve early diabetes diagnosis and care (16, 17). Natural language processing (NLP) enables computers to understand and generate human language. It is used in tasks such as speech recognition, language translation, sentiment analysis, and text summarization. Subcategories include syntax analysis, semantics analysis, named entity identification, machine translation, and question answering (18, 19). Natural language processing models, like ChatGPT, find diverse applications in medical sciences, covering diagnostics, research, treatment, decision-making, and scholarly writing. However, the reliability of ChatGPT in scientific writing is questionable due to its potential for generating unreliable references (20, 21).
1.3. Artificial Intelligence in Type 2 Diabetes Mellitus
Artificial intelligence has significant potential to enhance healthcare for diabetic patients. Machine learning, particularly in the context of T2DM, aids in early diagnosis, assisting both doctors and patients (22). Researchers have extensively explored various ML models to identify risk factors associated with the disease (23, 24).
2. Objectives
This study reviews the effectiveness of various ML models in achieving improved diagnostic accuracy and risk factor identification. As research in this field continues to evolve, we can expect even more sophisticated ML approaches to emerge, paving the way for a future of preventative T2DM care.
3. Methods
A literature review was conducted using the PubMed, Scopus, and Web of Science databases. These reliable tools were chosen due to their extensive healthcare-related content. The review focused on English-language documents published between 2019 and 2024. To curate relevant studies, we employed a comprehensive search query comprising the following keywords:
Diabetes, Diabetes mellitus, DM, Diabetic, T2DM, T1DM, Artificial intelligence, AI, Deep learning, Machine learning, Computational intelligence, Data mining, Pattern recognition, Neural network, Reinforcement learning, Diagnosis*, Identify, Detect*.
The inclusion and exclusion criteria for selecting the appropriate papers are mentioned in Table 1.
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Last 5 years | Duplicate articles |
| English articles | Studies with incomplete data or unannounced outcome |
| Research done on diabetes mellitus patients | Unrelated study designs including literature |
| Cohort or case-control studies | Review, case reports, book chapters |
| Related to the applications of artificial intelligence in the diagnosis of diabetes mellitus in patients | Studies with no access to their full text or abstract |
The Inclusion and Exclusion Criteria Used for Screening the Gathered Articles
3.1. Performance Metrics
Algorithm transparency and clinical assessment are crucial for vulnerable populations. Reported performance metrics varied across studies, with accuracy, F1 score, and AUC commonly employed for model evaluation.
Accuracy measures the proportion of correct predictions by a model. However, it can be misleading for imbalanced datasets, where predicting all observations as the majority class can inflate the accuracy score (25).
The F1 score is a machine-learning metric that combines precision and recall. It assesses a model’s accuracy by considering its class-wise performance rather than its overall performance. Precision measures how many of the predicted positive instances were actually positive. Recall measures how many of the actual positive instances were correctly predicted. The F1 score is the harmonic mean of precision and recall, ranging from 0 to 1, with a score of 1 representing the best possible performance (26).
Area under the curve (AUC) assesses model performance across various thresholds. It is applicable to classifiers providing confidence scores or probabilities. A perfect model achieves an AUC of 1, while a model with no discriminatory power results in an AUC of 0.5 (26).
4. Results
4.1. Analysis of Artificial Intelligence Models for Diagnosis
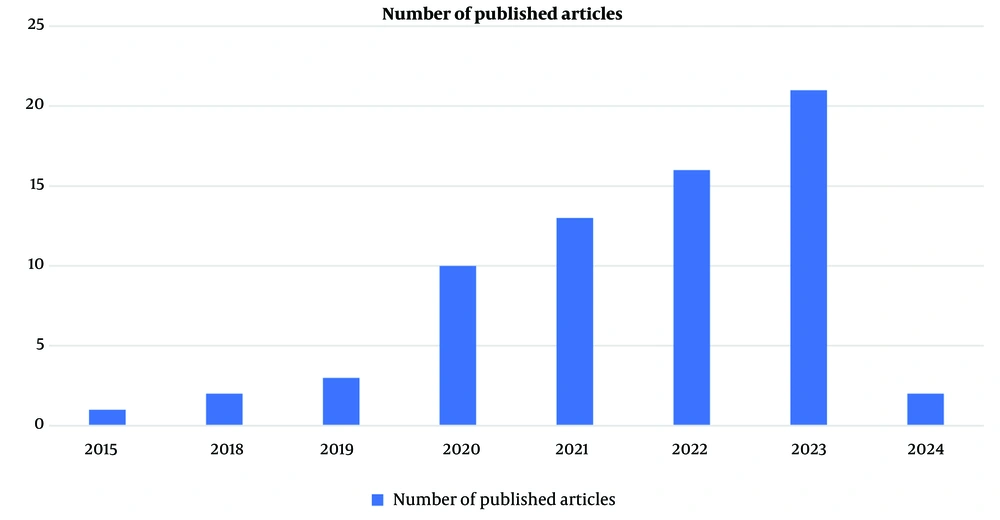
Among the 68 articles on AI-based diagnosis, 32 lacked full-text access and were excluded. The remaining 36 studies explored a variety of ML and DL models. Figure 1 highlights the growing interest in AI for diabetes diagnosis.
From these 68 articles, only 36 had access to full text, and 32 were discarded from this study.
Table 2 presents the details of the included studies.
| Author | Publication Year | Sample Size | Selected Features | AI Model | Model Algorithm | F1score | AUC | Accuracy |
|---|---|---|---|---|---|---|---|---|
| Albahli, S. (27) | 2020 | 253, 395 | FBS, HbA1c, gamma-GTP, BMI, TG, age, uric acid, sex, physical activity, drinking, smoking, and family history | ML | K-mean clustering + LR | Unmentioned | Unmentioned | 0.9753 |
| Eyasu, K. et al. (28) | 2020 | 12 | Unmentioned | NLP (Data mining) | J48 | 0.95 | Unmentioned | 0.9515 |
| PART | 0.944 | Unmentioned | 0.9451 | |||||
| JRip | 0.947 | Unmentioned | 0.9473 | |||||
| Islam, M. et al. (29) | 2020 | 1570 | Type of place, electricity, wealth index, age, education, working status, smoking, arm circumference, taking medicine, weight, and BMI | ML | SVM | Unmentioned | 0.662 | 0.929 |
| RF | Unmentioned | 0.593 | 0.923 | |||||
| Linear discriminant analysis | Unmentioned | 0.66 | 0.926 | |||||
| LR | Unmentioned | 0.682 | 0.925 | |||||
| NN | Unmentioned | 0.68 | 0.928 | |||||
| Bagged classification and regression tree (Bagged CART) | Unmentioned | 0.6 | 0.943 | |||||
| Kopitar, L. et al. (30) | 2020 | 3723 | A set of 58 variables that were not mentioned specifically. Generally, it includes an INDRISC (FR) questionnaire, physical activity (at least 30 min during the day), fruit and vegetable consumption, a history of antihypertensive drug treatment, a history of high blood glucose levels, and a family history of diabetes. | ML | Linear regression | Unmentioned | 0.854 | Unmentioned |
| Regularised generalised linear model (Glmnet) | Unmentioned | 0.859 | Unmentioned | |||||
| RF | Unmentioned | 0.852 | Unmentioned | |||||
| eXtreme gradient boosting (XGBoost) | Unmentioned | 0.844 | Unmentioned | |||||
| Light gradient boosting machine (lightGBM) | Unmentioned | 0.847 | Unmentioned | |||||
| Li, Y. et al. (31) | 2020 | 147 | Unmentioned | ML | SVM | Unmentioned | Unmentioned | 0.9722 |
| Liu, Y. (32) | 2020 | 650 groups | FBS, 2-hpp, clinical symptoms: Thirst, dry mouth, excessive drinking, polyphagia, polyuria, weight loss, family history, smoking and drinking | ML | MATLAB Neural Network | Unmentioned | Unmentioned | 0.92 |
| Al Masud, F. et al. (33) | 2021 | 306 | Age, area of residence, standard growth rate, HbA1c, hypoglycemia, adequate nutrition, autoantibodies, sex, and family history of type 1 and 2 diabetes | ML | Ranker analysis, data mining | Unmentioned | Unmentioned | Unmentioned |
| Deepa, S.N and Banerjee, Abhik (34) | 2021 | 900 | Images of the tongue | ML | CNN + SVM | 0.9831 | Unmentioned | 0.9782 |
| Dietz, B. et al. (35) | 2021 | 2371 | T1-weighted whole-body MRI, sex, age, BMI, insulin sensitivity, and HbA1c | DL | Dense CNN | Unmentioned | 0.87 | Unmentioned |
| Islam, M. et al. (36) | 2021 | 492 | Retinal images | DL | CNN | Unmentioned | 0.662 | Unmentioned |
| Lee, W.S. et al. (37) | 2021 | 1000 | Synthetic glucose profiles | ML | Shallow neural network | Unmentioned | Unmentioned | 0.873 |
| DL | Multilayer perceptron (MLP) | Unmentioned | Unmentioned | 0.9 | ||||
| CNN | Unmentioned | Unmentioned | 0.865 | |||||
| RNN | Unmentioned | Unmentioned | 0.0866 | |||||
| Samreen, S. (38) | 2021 | 520 | Age, sex, polyuria, polydipsia, sudden weight loss, weakness, polyphagia, genital thrush, visual blurring, itching, irritability, delayed healing, partial paresis, muscle stiffness, alopecia, and obesity | ML | NB | Unmentioned | 0.95 | 0.8961 |
| KNN | Unmentioned | 0.98 | 0.9487 | |||||
| LR | Unmentioned | 0.97 | 0.9269 | |||||
| SVM | Unmentioned | 0.99 | 0.9833 | |||||
| DT | Unmentioned | 0.96 | 0.9685 | |||||
| RF | Unmentioned | 0.99 | 0.9833 | |||||
| Adaboost(AB) | Unmentioned | 0.98 | 0.9641 | |||||
| Gradient boost (GB) | Unmentioned | 0.99 | 0.9717 | |||||
| Srivastava A.K, et al. (39) | 2021 | Unmentioned. Pima Indian diabetes dataset | Unmentioned | ML | DNN | 0.8931 | 0.9236 | 0.9474 |
| Xiang, Y. et al. (40) | 2021 | 165 | Fundus images, tongue appearance, and pulse characteristics | ML | RF | 0.76 | Unmentioned | 0.85 |
| Zhang,k. et al. (41) | 2021 | 57672 cases and 115344 retinal images | Fundus images, age, sex, height, weight, BMI, and blood pressure | DL | RF | Unmentioned | Unmentioned | 0.93 |
| ML | CNN | Unmentioned | Unmentioned | 0.861 | ||||
| Alshari, H. and Odabas, A. (42) | 2022 | 14682 | Physical activity, dietary, smoking features, alcohol consumption, hypertension, age, gender, race, marital status, education level, annual family income, and the ratio of family income to poverty guidelines | ML | XGBoost | 0.748 | 0.842 | 0.846 |
| LightGBM | 0.749 | 0.843 | 0.846 | |||||
| CatBoost | 0.737 | 0.836 | 0.836 | |||||
| Neural networks | 0.721 | 0.821 | 0.829 | |||||
| Anaya-Isaza, A. and Zequera-Diaz, M (43) | 2022 | 167 | Foot thermography | DL | CNN | 0.8583 | Unmentioned | 0.8278 |
| Balasubramaniyan, S. et al. (44) | 2022 | 2675 | Tongue images: Tongue shape and color, fissure identification, fur color and fur thickness, tooth markings, and red dot | DL | CNN | 0.99 | Unmentioned | 0.984 |
| Ellouze, A. et al. (45) | 2022 | 768 | Pregnancy, plasma glucose concentration, diastolic blood pressure, triceps skinfold thickness, insulin, mass, pedigree of diabetes, and age | ML | KNN | 0.77 | 0.76 | 0.77 |
| SVM | 0.8 | 0.87 | 0.8 | |||||
| DT | 0.76 | 0.79 | 0.75 | |||||
| DL | RNN | 0.93 | 0.95 | 0.93 | ||||
| CNN | 0.9 | 0.92 | 0.9 | |||||
| Long short-term memory (LSTM) | 0.97 | 0.99 | 0.97 | |||||
| Long short-term memory (LSTM) | 0.95 | 0.97 | 0.95 | |||||
| Fufurin, I. et al. (46) | 2022 | 1200 infrared exhaled breath spectra from 120 volunteers | Exhaled breath samples and using IR breath spectra | DL | CNN | Unmentioned | 0.99 | 0.997 |
| Hossain, E. et al. (47) | 2022 | Unmentioned | Number of pregnancies, BMI, insulin levels, age, glucose, skin fold thickness, blood pressure, and diabetes pedigree function | ML | KNN & LightGBM | 0.84 | 0.936 | 0.891 |
| Rabie, O. et al. (48) | 2022 | 829 | Age, BMI, glucose, the number of pregnancies, blood pressure, skin foldthickness, two-hour insulin, and pedigree diabetes function | DL | DNN | 0.92 | Unmentioned | 0.9307 |
| Ullah, Z. et al. (49) | 2022 | 253680 | Comprised a total of 22 features: Blood pressure, high cholesterol, BMI, smoking, Stroke, heart diseases, fruits, veggies, heavy alcohol consumption, any health care, sex, age, etc. | ML | Nearest neighbor (SMOTE-ENN) | 0.98 | 0.98 | 0.9838 |
| Zee, B. et al. (50) | 2022 | 2221 | Retinal imaging with a non-mydriatic fundus camera | ML | SVM | Unmentioned | 0.993 | Unmentioned |
| Garcia-Dominguez, A et al. (51) | 2023 | 1019 | Diastolic blood pressure, systolic blood pressure, glucose, height, LDL, waist circumference, TG, education, insulin, gender, cholesterol, and age | ML | Neural network | 0.86 | 0.934 | 0.86 |
| Iparraguirre-Villanueva, O.et al. (52) | 2023 | 768 | Number of pregnancies, glucose level, diastolic blood pressure, thickness of skin folds, insulin levels, BMI, genetic history of diabetes, and age | ML | Nearest neighbor(NN) | Unmentioned | 0.667 | 0.753 |
| Naïve Bayes(NB) | Unmentioned | 0.677 | 0.461 | |||||
| Decision tree(DT) | Unmentioned | 0.602 | 0.708 | |||||
| LR | Unmentioned | 0.555 | 0.698 | |||||
| SVM | Unmentioned | 0.56 | 0.67 | |||||
| Salem Alzboon, M. et al. (53) | 2023 | 768 | Data set of 8 demographics and clinical details: Age, gender, number of pregnancie, BMI, blood pressure, skin thickness, insulin level, and glucose concentration | ML | LR | 0.613 | 0.828 | Unmentioned |
| DT | 0.567 | 0.665 | Unmentioned | |||||
| RF | 0.576 | 0.811 | Unmentioned | |||||
| KNN | 0.56 | 0.776 | Unmentioned | |||||
| NB | 0.64 | 0.808 | Unmentioned | |||||
| SVM | 0.583 | 0.822 | Unmentioned | |||||
| GB | 0.528 | 0.636 | Unmentioned | |||||
| Neural network | 0.61 | 0.825 | Unmentioned | |||||
| Deepa,K. and Ranjeeth Kumar, C .(54) | 2023 | Unmentioned | Unmentioned | ML | DT | Unmentioned | Unmentioned | 0.77 |
| KNN | Unmentioned | Unmentioned | 0.773 | |||||
| LR | Unmentioned | Unmentioned | 0.793 | |||||
| Ensemble method | Unmentioned | Unmentioned | 0.806 | |||||
| Duc, L. et al. (55) | 2023 | Unmentioned | Unmentioned | ML | SVM + ANN | Unmentioned | 0.96 | 0.938 |
| Nguyen et al. (56) | 2023 | 2153 | Gender, age, MI, waist circumference, hip circumference, systolic blood pressure, diastolic blood pressure, FBS, 2-hPP, total cholesterol, TG, HDL and insulin | ML | RF | 0.94 | 0.94 | 0.85 |
| Nilashi, M. et al. (57) | 2023 | 768 | Number of pregnancy, 2-hPP, diastolic blood pressure, triceps skin fold thickness, 2 hours serum insulin, BMI, diabetes pedigree function, and age | DL | DBN | Unmentioned | Unmentioned | 0.9832 |
| Önal et al. (58) | 2023 | 68 | Full irtis images, the iris segmentation from raw images, the segmentation of the pancreatic region in the iridology chart | DL | CNN | 0.8333 | Unmentioned | 0.8 |
| Shaukat, Z.et al. (59) | 2023 | 768 | Number of pregnancies, plasma glucose concentration,diastolic blood pressure,triceps skinfold thickness,2-hour serum insulin,BMI,diabetes pedigree function, and age | ML | DT | 0.72 | 0.839 | 0.7186 |
| KNN | 0.72 | 0.697 | 0.723 | |||||
| RF | 0.78 | 0.832 | 0.7879 | |||||
| LR | Unmentioned | 0.848 | 0.7966 | |||||
| SVM | 0.79 | 0.723 | 0.7922 | |||||
| Zhang, J. et al. (60) | 2023 | 820 | A set of nine clinical feature: Admission glucose, BMI > 28, cardiovascular disease, age, NAFLD, ALT, HDL-C < 1.03, UA, and smoking | ML | LR | 0.357 | 0.819 | Unmentioned |
Delves Deep into the Effectiveness of Various Machine Learning (ML) and Deep Learning (DL) Algorithms for Diagnosing Diabetes Mellitus
4.2. Machine Learning Techniques
4.2.1. Support Vector Machines (SVMs)
support vector machine is an ML method primarily used to classify data by finding a maximum decision boundary to separate data from different classes. The strengths of SVMs are their ability to handle high-dimensional features and various types of unstructured data, such as text and images. However, SVMs struggle when classes overlap significantly and when high dimensionality leads to overfitting, resulting in poor generalization to new data (61).
SVM performance is promising, with studies achieving high accuracies. For instance, Wang et al. employed SVMs with radial basis function (RBF) kernels and achieved accuracies exceeding 90%, highlighting the importance of proper kernel selection in model performance (62). Similarly, Ellouze et al. reported 80% accuracy, while Zee achieved an exceptional 99.3% accuracy by using internal and external validation, demonstrating potential for practical applicability (45, 50).
However, Iparraguirre-Villanueva et al. observed a lower accuracy of 56%, highlighting potential limitations with specific datasets or parameter tuning (52). Further investigation into SVM kernel selection, optimization techniques, and generalizability using larger and balanced datasets alongside adequate validation could be beneficial (Appendix 1).
4.2.2. Random Forests (RFs)
Random forest is an ML method that combines multiple decision tree outcomes to report the best single result. Similar to SVM, RF can be used in classification and regression tasks and can handle high-dimensional spaces. However, the complexity of RFs in generating explanations decreases their interpretability (63). These models consistently demonstrate strong performance. Islam et al. achieved an outstanding F1 score of 0.99, signifying excellent classification ability (36). Similarly, Nguyen et al. reported an accuracy of 85% and an AUC of 0.94, showcasing RFs' capability for accurate diabetic/non-diabetic case identification (56). Studies by Shaukat et al. (accuracy: 78.79%) and Xiang et al. (accuracy: 85%) suggest that RFs might benefit from dataset-specific optimization for peak performance (40, 59). Although increasing the number and depth of trees by using larger datasets can improve the algorithm's robustness, it should be considered that it leads to longer training times and requires more memory than other algorithms (63). See Appendix 2 for detailed results.
4.2.3. K-Nearest Neighbors (KNNs)
This ML method finds the most probable prediction by grouping the inputs. The inputs that have the least distance on the chart are gathered in a group. The value of the group is determined by the mean or the majority vote of each group participant. . K-nearest neighbors's simplicity and ease of implementation make it a good choice for real-world applications, such as in the medical field, where understanding the decision-making process is essential for trusting the model outcomes. However, its simplicity struggles to handle large, imbalanced, and complex data (45).
K-nearest neighbor results show considerable variation. Samreen reported a high accuracy of 94.87%, and Ellouze et al. achieved a moderate accuracy of 77% (38, 45). Conversely, Iparraguirre-Villanueva et al. observed a lower accuracy of 66.7% (52) (Figure 2). This variability emphasizes the sensitivity of KNNs to data characteristics and parameter selection. Further research could explore techniques for optimal K-value selection and distance metric choice for enhanced performance. See Appendix 3 for detailed results.
4.2.4. Logistic Regression
Logistic regression (LR) is a discriminative model for binary classification that models the probability of an input belonging to a class using a logistic function. It employs coefficients to predict probabilities. Logistic regression is a straightforward algorithm, making it accessible to apply, train, and interpret. However, it assumes linear relationships between variables and log odds of outcomes, which can reduce its flexibility and suitability for real-world data (57, 64).
Logistic regression yielded mixed results. Islam et al. achieved an accuracy of 66.2% using LR, whereas Salem Alzboon et al. reported an accuracy of 61.3% and an AUC of 0.828 (29, 53). However, Villanueva et al. documented a lower accuracy of 55.5% for LR (52). These findings suggest limitations of LR in specific scenarios, potentially due to dataset linearity assumptions. Exploring regularization techniques and feature engineering could be beneficial for improving LR's effectiveness in diagnosing diabetes. See Appendix 4 for detailed results.
4.2.5. Neural Networks (NNs)
Neural networks predict the outcome of interest by passing input information through layers of interconnected neurons with learnable weights. Each neuron applies an activation function to the weighted sum of inputs, enabling complex nonlinear mappings. Through forward and backward propagation, NNs learn to minimize a loss function, adjusting weights to generate accurate predictions (15).
The performance of NNs varied considerably. Liu achieved an accuracy of 92% using a MATLAB Neural Network, while Salem Alzboon et al. reported an accuracy of 61% for a general neural network (32, 53). Garcia-Dominguez et al. documented a moderate accuracy of 86% for a neural network (51). These variations highlight the dependence of NN performance on factors like network architecture, training data size and quality, and hyperparameter tuning. Studies by Srivastava et al. (accuracy: 89.31%) and Rabie et al. (accuracy: 92%) further showcase the potential of NNs with careful optimization (39, 48). See Appendix 5 for detailed results.
4.3. Deep Learning Techniques
Deep learning approaches, including CNNs and RNNs, demonstrated promising results in some studies. Their ability to automatically learn from data without manual feature extraction and labeling makes these techniques suitable for tasks where defining features is challenging, such as medical image processing. However, clinical applicability remains difficult due to the complexity of interpreting and identifying errors or biases in the models (16, 17).
Anaya-Isaza and Zequera-Diaz achieved an accuracy of 85.83% using CNNs, while Ellouze et al. reported an accuracy of 97% for LSTMs (a type of RNN) (43, 45). Conversely, Önal et al. documented a lower accuracy of 83.33% for a CNN, suggesting that DL techniques might require substantial data for optimal performance (58). Further research into data augmentation techniques and transfer learning could be beneficial for improving the generalizability of DL models in diabetes diagnosis. See Appendix 6 for detailed results.
4.4. Ensemble Methods
A limited number of studies explored ensemble methods. Duc et al. combined SVM and an artificial neural network, achieving an accuracy of 93.8% (55). Deepa and Ranjeeth Kumar reported an accuracy of 80.6% for an ensemble method, indicating their potential effectiveness in diabetes diagnosis (54). Further investigation into ensemble methods that combine the strengths of different algorithms is warranted.
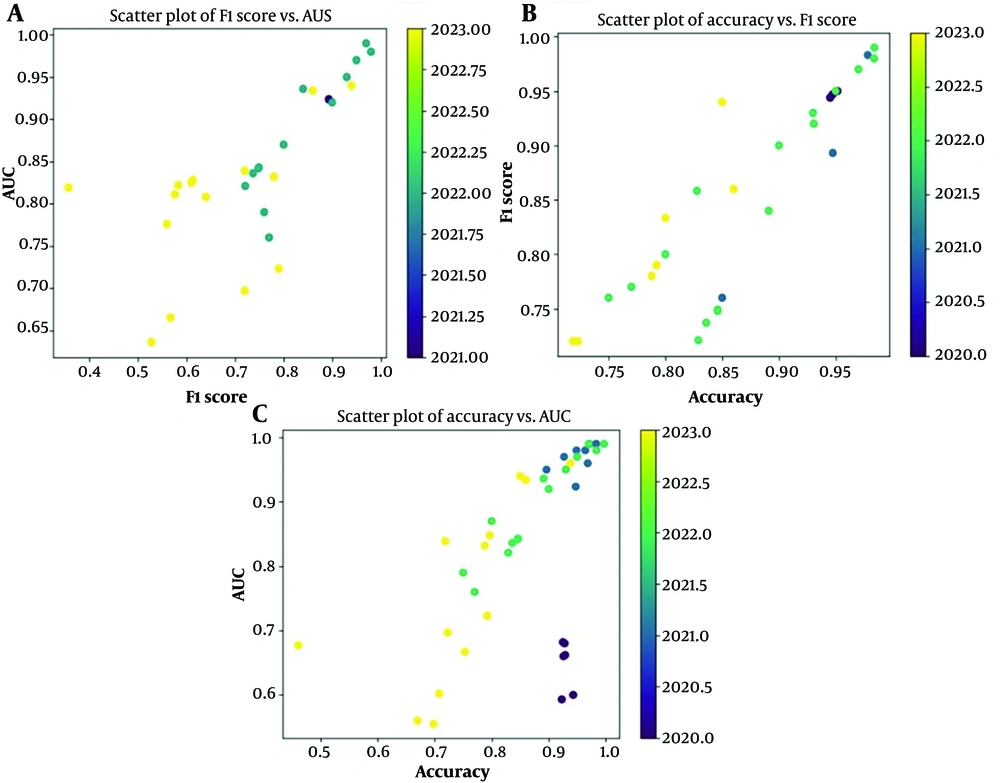
Figure 3 presents three scatter plots that analyze the performance of various AI models designed to diagnose DM at an early stage. These plots utilize three key metrics: Area Under the Curve, F1 score, and accuracy. The upper left scatter plot depicts the relationship between F1 score and AUC. Here, we observe that all the models achieve relatively good performance, with F1 and AUC scores consistently exceeding 0.65. The upper right scatter plot focuses on the models' accuracy compared to their F1 score. Interestingly, most data points reside above the diagonal line, signifying that overall accuracy tends to be higher than the F1 score. This implies that the models excel at correctly classifying the majority of cases but might struggle with specific subgroups within the data. For example, a model might be adept at identifying healthy individuals yet less proficient at distinguishing between pre-diabetic and diabetic patients. The lower scatter plot explores the models' performance using both AUC and accuracy. This plot reveals a positive finding: A cluster of data points occupies the upper right quadrant. This positioning signifies that a substantial portion of the models exhibit both high AUC and high accuracy, effectively differentiating between individuals with and without DM. Notably, a few models stand out in the upper right corner, achieving exceptionally high values for both metrics.
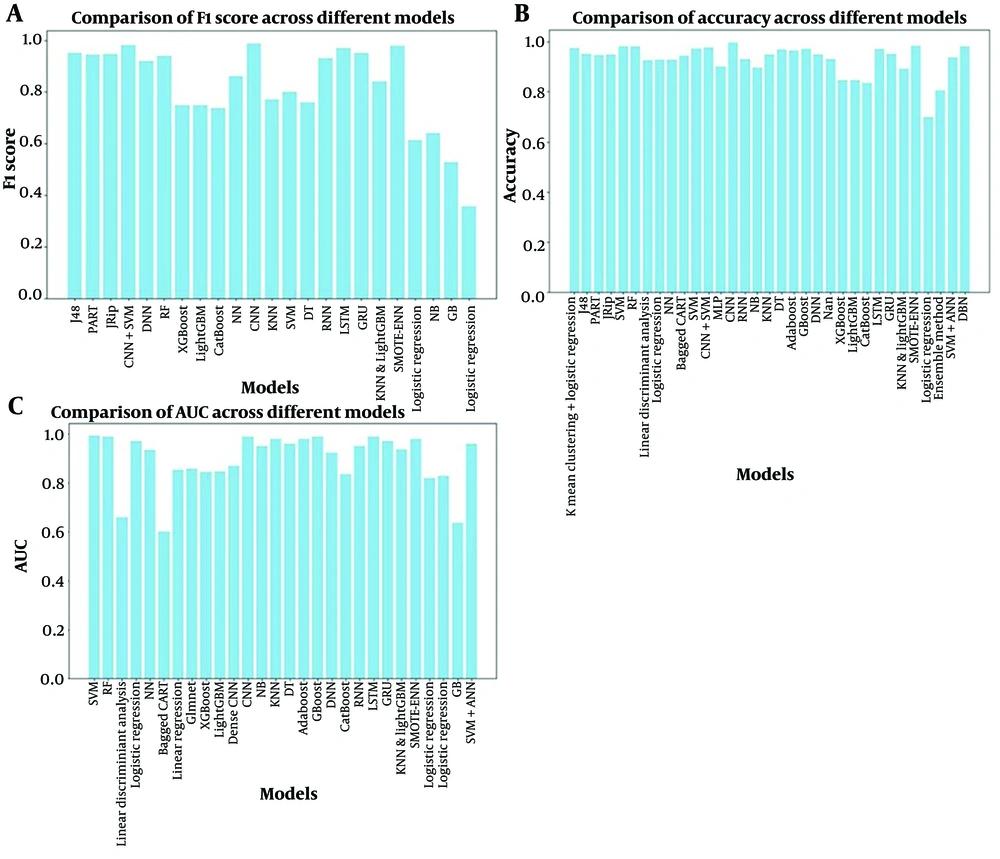
Figure 4 presents a comparative analysis of the algorithms implemented within various ML models. The evaluation emphasizes the F1 score and AUC as the primary performance metrics. This choice is driven by the critical need to accurately identify pre-diabetic and diabetic individuals within a population. Notably, the results demonstrate that many models achieve F1 scores and AUCs exceeding 80%. This high performance suggests significant promise for the real-world application of these AI models in clinical settings. Their ability to accurately identify pre-diabetic and diabetic patients can significantly improve preventative measures and patient outcomes.
The F1 score is a metric that balances precision and recall. Area Under the Curve represents the probability that the model will rank a positive instance higher than a negative instance. Accuracy is the proportion of correct predictions made by the model.
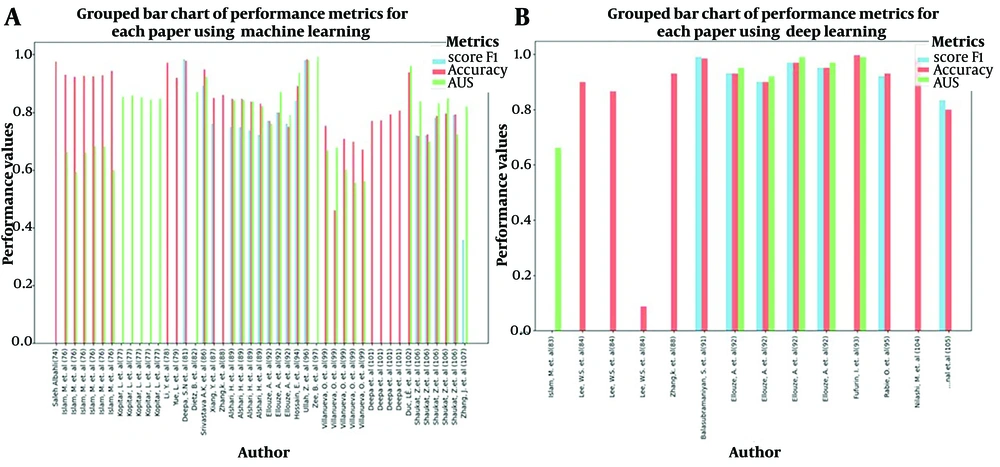
Figure 2 shows a grouped bar chart comparing the performance metrics of various research papers. Each bar group represents a paper by a specific author, and the bars within each group represent performance on three metrics: F1 score, accuracy, and AUC. These metrics are color-coded for easy differentiation.
5. Discussion
In summary, AI, particularly ML and DL, can revolutionize diabetes management. Machine learning methods (SVMs, RFs) excel in diagnosis, while CNNs show promise.
Multiple publications have addressed the topic of our review, exploring AI applications in DM diagnosis to various extents. A significant number of them aimed to review studies with ML algorithms to screen complications of DM, such as cardiovascular complications and diabetic retinopathy (65). As mentioned earlier, DL models, especially CNNs, are widely used for medical imaging recognition and have shown promising outcomes. Most of these studies used CNN-based models trained on multi-center medical imaging datasets or public datasets, such as the Indian Diabetic Retinopathy Image Dataset (66). Other papers have investigated the potential of diabetes prediction and early detection to initiate timely intervention and improve outcomes by utilizing ML-based models. Since there is a broad spectrum of risk factors, symptoms, and signs for this disorder, various features are used in each article (67). Some of these models were trained on data extracted from electronic medical records as a large, real-time clinical data source (68). Much effort has been made to identify the most weighted demographic, clinical, or laboratory features for developing a predictive model with maximum accuracy. Parameters such as blood sugar level, BMI, triglyceride level, HbA1c, and pregnancy have showcased the most predictive value (69). Our study is not limited to a particular diagnostic method, as we have compared various AI-powered advancements in the field of DM diagnosis. According to our results, the accuracy of the model increased when DL and ML techniques were used simultaneously. However, there are challenges to overcome. The lack of sufficient data, especially for specific groups or rare types of diabetes, can hinder the effective use of AI. Understanding complex AI models, particularly DL ones, is a concern for healthcare providers who need to understand why the models make certain predictions. Additionally, the lack of standard methods to measure AI performance makes it difficult to compare different studies.
To maximize the benefits of AI in diabetes care, we need to address these challenges. Gathering more data while protecting patient privacy is essential. Research on making AI models more understandable, such as using explainable AI (XAI), can help build trust and facilitate the broader adoption of AI in healthcare.
5.1. Future Research Directions and Open Challenges
Advancing AI in DM diagnosis and addressing evolving challenges requires further research in several key areas. The following research directions can help improve the efficiency and reliability of AI systems in this field.
First, developing sustainable AI models necessitates the availability of rich and high-quality data. Future studies should focus on expanding datasets to include a broader range of populations and clinical scenarios. Additionally, the collection of longitudinal data and real-world evidence can enhance the generalizability of AI algorithms in diabetes diagnosis (70).
Second, researchers should explore methods to identify and mitigate biases and errors in diabetes diagnosis algorithms. Promoting transparency in algorithm development and decision-making processes can build trust among stakeholders. Moreover, the interpretation of AI models is crucial for clinical acceptance. Future research should concentrate on developing explanatory AI techniques that provide insights into how the model makes its predictions. By improving the interpretability of the model, doctors can better understand and trust the recommendations of AI systems when diagnosing diabetes (71). Third, to facilitate the adoption of AI tools in clinical practice, future research should explore ways to seamlessly integrate these systems into existing healthcare workflows. Collaboration between AI researchers and healthcare providers can help adapt AI solutions to the specific needs and constraints of the clinical environment, ultimately improving the utility and impact of AI in diabetes diagnosis. Pursuing these lines of research can advance the field of AI in diagnosing diabetes, ultimately improving patient outcomes and healthcare (72).