1. Background
Breast cancer is the second cause of death worldwide and the most invasive cancer amongst women. The main risk factor in breast cancer patients is metastasis. Increasing evidence indicates that CD44 promotes metastasis in multiple cancer types, such as breast, bladder, pancreatic, prostate, and ovarian cancer (1). Several studies showed that the rate of extravasation and metastasis is raised in aggressive breast cancer with a high level of CD44 or more CD44+ circulating tumor cells in the blood. This phenomenon is also associated with increased recurrence, leading to decreased disease-free survival rate in patients (2). CD44 is the main receptor for glycosaminoglycan hyaluronic acid (HA), an adhesion/homing molecule, which is universally expressed in physiological and pathological conditions that contributes to major oncogenic signaling networks, by stimulating every feature of tumor progression (3). Decades of research have shown that aggressive breast carcinoma cells, in addition to synthesizing high levels of HA, also have a high level of CD44 expression. Hyaluronic acid, as the main constituent of the tumor extracellular matrix (ECM), accumulates at sites of cell division and is involved in fast matrix remodeling. This process is followed by inflammation, embryonic morphogenesis and tumorigenesis, leading to metastasis and cancer invasion (4). HA not only provides a hydrophilic matrix and extracellular support but also regulates cell-cell adhesion, growth, differentiation, and cell migration (5) by directly attaching to HA receptors such as lymphatic vessel endothelial hyaluronan receptor (LYVE-1), CD44 receptor for hyaluronan-mediated motility (RHAMM), Toll-like receptor 4 (TLR4), or by stimulating other proteins (6).
Lately, the role of EMT in the pathogenesis of metastasis and fibrosis of carcinogenic tumors has been defined. The event of tumor metastasis mediated by EMTs, often facilitated by snail/slug as an inducer of EMT, directly inhibits epithelial markers such as E-cadherin (a gatekeeper of the epithelial phenotype) by up-regulating mesenchymal markers (6). Moreover, a low level of E-cadherin reduces the cell-cell adhesion and facilitates metastatic behavior and invasive phenotype of carcinomas. Previous studies showed cells that express lower levels of E-cadherin, exhibit increased binding affinity between CD44 and HA (7). It was recently shown that CD44-HA signaling stimulates invasion by triggering Extracellular Signal-regulated Kinase-1 (Erk) pathways, growth factor receptor signaling phosphatidylinositol 3-kinase (PI3K/Akt), and NFkB signaling, which can individually induce EMT via snail expression (8). Owing to the critical role of HA/CD44 interactions in cancer development and metastasis, lately, inhibition of this interaction was used as a potential target for cancer therapy. There are several methods to disrupt the CD44/HA interaction such as inhibition of HA/CD44 interactions by CD44 siRNAs (9), small hyaluronan oligosaccharides (10), overexpression of soluble CD44, and the use of blocking antibody against HA binding site of CD44 (11). Among these platforms, aptamers (single-stranded RNA or DNA oligonucleotides that can bind specifically to the target, using their tertiary structure) have attracted much attention due to their small size (usually from 20 to 60 nucleotides), high- affinity binding to a target, and specificity.
2. Objectives
Based on the evidence, we hypothesized that there is a positive relationship between CD44/HA signaling and snail expression. Therefore, to better understanding CD44/HA interaction and the associated effect on snail expression and invasive behavior, we disrupted CD44/HA interaction by DNA aptamer that specifically bind to the HABD of CD44. Then, we evaluated its treatment effect on snail expression and invasion of metastatic breast cancer cell lines, MDA-MB-231, cultured in 2D and 3D, which is a more in vivo-like systems.
3. Methods
3.1. Selection of Aptamers
The aptamer was selected based on Somasunderam et al. (5) method with some modifications. In the Somasunderam protocol, a thioaptamer against CD44 was designed. Thioaptamers are modified aptamers with the sulfur substitution of the phosphate backbone. This modification improves the binding affinity of the thioaptamer to the target protein and also increases its non-specific binding to other proteins. Here, we selected and synthesized aptamer sequence (Metabion Company, Germany) with a phosphate backbone.
3.2. 2D Culture
The human breast cancer cell lines MDA-MB 231 and SKBR3 were purchased from the Pasteur Institute of Iran. Approximately, 5 × 105 cells from each cell lines were seeded in a 6-well plate, in RPMI-1640 medium, supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc., Waltham, MA, USA). After incubation for 24 h, the aptamer was added to the cells at a density of 50 ng.mL-1 (synthesized by Metabion Company, Germany), followed by treatment with a combination of aptamer and Hyaluronan (Low Molecular Weight, manufactured by Lifecore Biomedical, LLC) for a period of 48 h.
3.3. Flow Cytometry
The cell surface binding capacity of the selected aptamer was determined by flow cytometry assay. In brief, 5 × 105 MDA-MB 231 and SKBR3 cells (as a negative control) were stained with FITC-conjugated aptamer and commercial antibody (Abcam, UK) (as a positive control). After three sequential washes with washing buffer (PBS 5% tween 20), the bound aptamer and antibody were measured by a FACS Calibur (BD Bioscience, USA).
3.4. Three-Dimensional Cell Culture Methods
MDA-MB-231 adherent cells were trypsinized, and 5x105 cells were divided into 1.5 mL polypropylene cone-shaped tubes and centrifuged for 5 min at 500 ×g. The cell pellet was incubated at 37°C, 5% CO2 for 24 h. The RPMI medium was supplemented with 10% FBS, and then restored with a medium containing HA (1 mg/mL) and aptamer 50 ng.mL-1. The medium replacement was performed every 24 h, and the aggregates were harvested after 48 h of incubation.
3.5. Quantitative Real-Time PCR
Quantitative real-time PCR was performed using an ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Twenty-five microliter reaction mixture was prepared as follows: 2 μL of cDNA (ten-fold dilution), 0.5 μL of each forward and reverse primers, with a concentration of 5 pmol/L, and 12.5 μL of SYBR green DNA PCR Master Mix, in a final volume of 25 μL, by adding nuclease-free water. After the initial denaturation for 10 min at 95°C, the amplification step was carried out for 40 cycles, denaturation at 95°C for 15 s, annealing and elongation at 60°C for 30 s. The final elongation time was 10 min at 72°C to permit to complete the reaction products. A melting curve was plotted from 70 cycles of 10 s each with a temperature increase of 0.5°C/cycle, initiating at 60°C. The slope of the standard curve represents the efficiency of the amplification. Data analysis was performed using the 7500 software V 2.0.1. The relative expression of the snail mRNA gene was calculated using the 2−ΔΔCT formula by comparing it with β-actin, as the internal control. The sequences of specific primers were used to detect snail listed below: Forward, 5’-GAGGCGGTGGCAGACTAG-3’ and reverse, 5’-GACACATCGGTCAGACCAG-3’. As the reference gene, the levels of β-actin in the samples were also evaluated. Primers for amplification of β-actin were as follows: β-actin forward, 5’-AATCGTGCGTGACATTAAG-3’ and reverse, 5’-GAAGGAAGGCTGGAAGAG-3’.
3.6. Preparation of RNA and cDNA Synthesis
Total RNA was extracted using BioZOL RNA extraction reagent (BioFlux Corporation, Tokyo, Japan), and its concentration was measured by a NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). To remove genomic DNA contamination, total extracted RNA was treated with an RNase-free DNaseI enzyme, according to the kit instruction (Promega, Madison, WI, USA). Total RNA was instantly reversed transcribed to first‑strand complementary DNA by adding random hexamer and oligo dT primers, using the revert aid first-strand cDNA synthesis kit, according to the manufacturer’s instructions (Fermentas, Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA).
3.7. Wound Healing Assay
To determine wound healing (cell motility) function, the cells were cultured in 6-well plates. After reaching the proper confluence, the cells were exposed to serum starvation for 24 h. A scratch wound was generated in the cell monolayer, using a 10 µL pipette tip. The cells were washed twice with PBS to eliminate the floating cells. The development of cell motility in the wound area was detected at different time periods (24 and 48 h). A photograph of cell movement was taken using a digital camera attached to a microscope (Olympus Corporation, Tokyo, Japan). The percentage of wound closure was determined, using OLYSIABioReport imaging software (Informer Technologies, Inc., Madrid, Spain). Each assay was carried out in triplicate.
3.8. Statistical Analysis
Data were collected and expressed as the mean ± standard error of the mean from three independent experiments. Statistical analysis was performed by applying one-way analysis of variance and Tukey’s test to compare the control and vehicle groups against the treated groups. P < 0.05 was considered statistically significant.
4. Results
4.1. Membrane Binding Capacity of FITC-Conjugated Aptamer to CD44 by Flow Cytometry
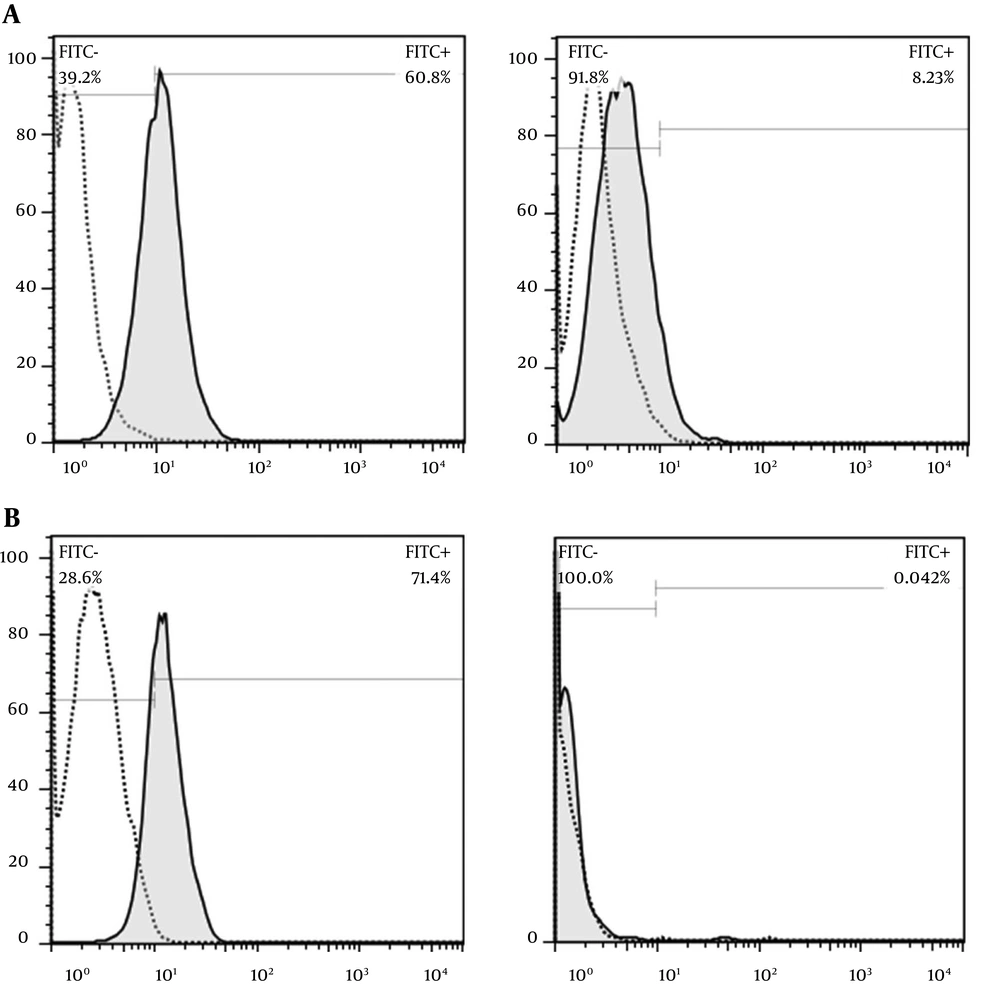
Investigation of binding affinity of the FITC-conjugated selected aptamer to cell surface receptor, CD44, revealed that the selected aptamer reacts 60% with CD44 on MDA-MB 231 cell line (CD44 positive cell line) and represents no non-specific reaction to SKB-R3 cell line (CD44 negative cell line). Evaluation of membrane binding CD44 antibody showed 70% reactivity with MDA-MB 231 cell line (Figure 1).
Investigation of membrane binding of CD44 compared to the CD44 antibody by flow cytometry. A, CD44 aptamer (100 nM) showed 60% reactivity with MDA-MB 231 and 8% nonspecific binding to SKB-R3 cell line. B, the membrane binding of the CD44 antibody to MDA-MB 231 cell line was 71% and no non-specific binding to SKB-R3 was found.
4.2. Effect of CD44 Aptamer and Its Combination with Hyaluronic Acid on Snail Expression
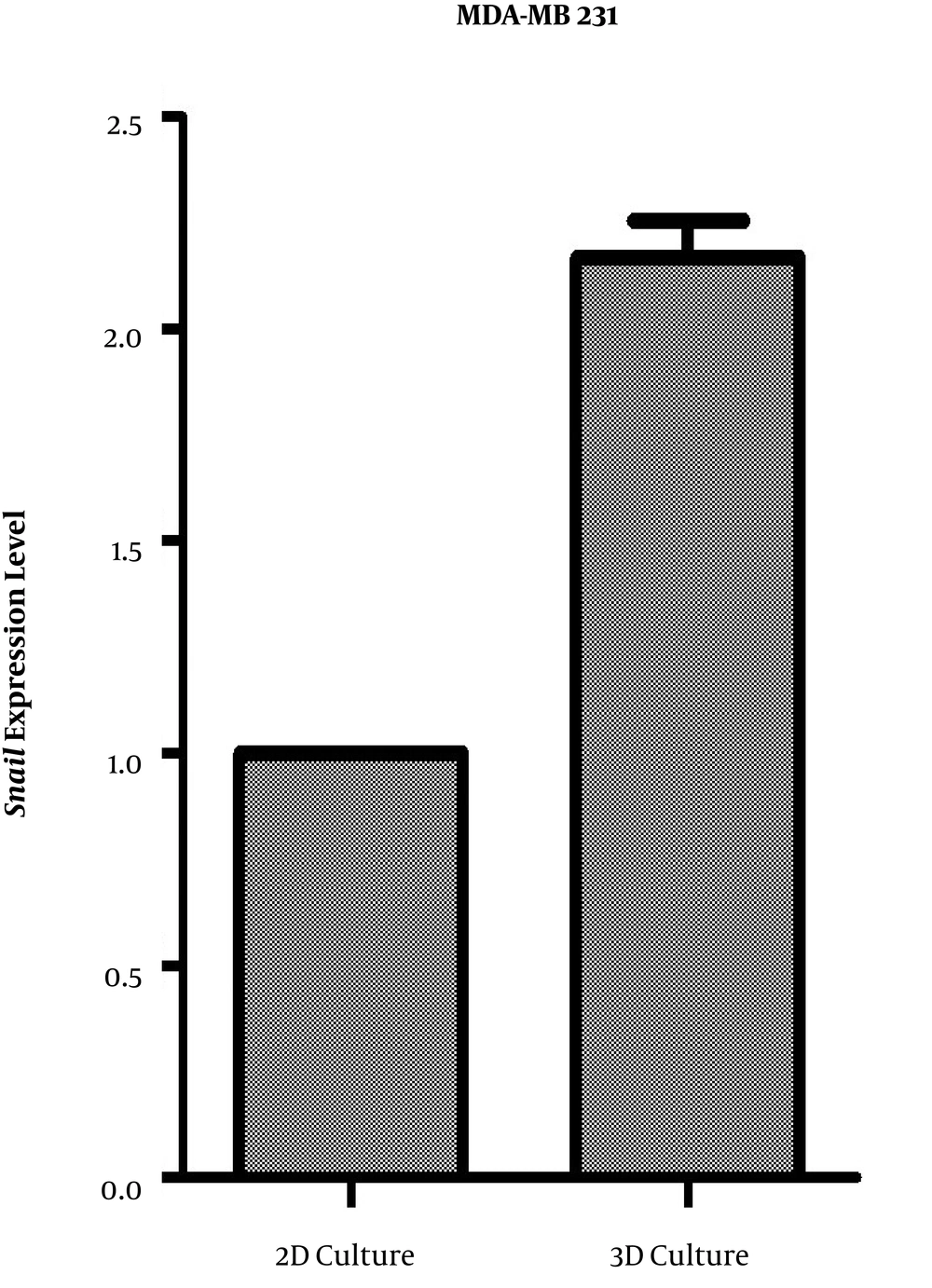
MDA-MB-231 cells were grown for 24 h before being treated with the CD44 aptamer, separately and in combination with hyaluronic acid in 2D and 3D culture. The 3D culture of MDA-MB-231 cells led to a lower increase in the snail mRNA than 2D cultured cells (Figure 2).
Quantification of snail mRNA expression by a quantitative real-time PCR in MDA-MB 231 cell line in 3D and 2D culture. Data are the relative expression of snail mRNA to those of β-actin in the treated groups compared to the control cells. Data are presented as mean ± SD of three independent experiments (*P < 0.05).
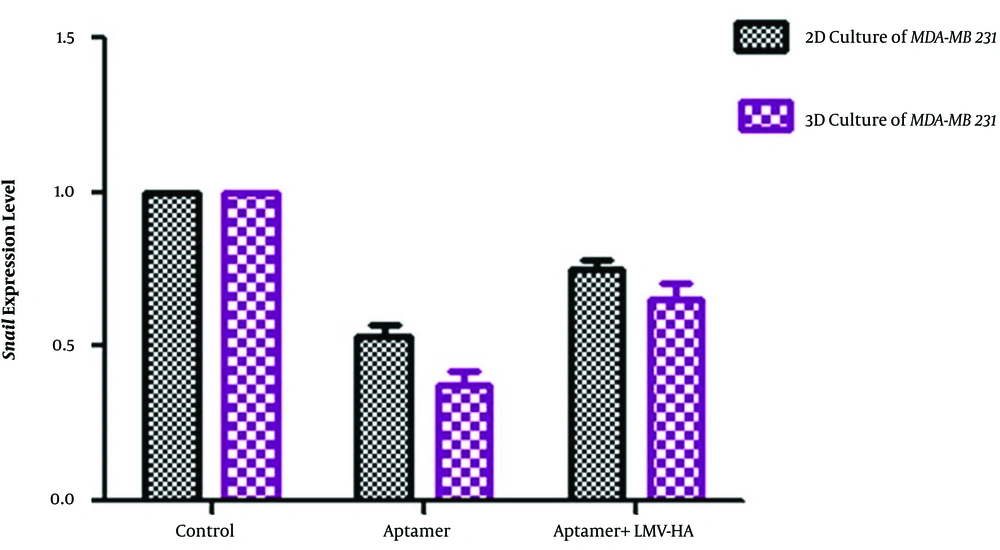
The data show that treating breast cancer cells with the CD44 aptamer, and its combination with hyaluronic acid can induce a significant reduction in the snail mRNA expression in both 2D and 3D cultures (Figure 3). The greatest significant reduction in snail mRNA expression was detected after incubating cells that were cultured in 3D condition with 50 nM of the CD44 aptamer.
Quantification of snail mRNA expression by a quantitative real-time PCR in MDA-MB 231 cell line treated with aptamer and aptamer + hyaluronic acid in 3D and 2D culture. Data are the relative expression of snail mRNA to those of β-actin in the treated groups compared to the control cells. Data are presented as mean ± SD of three independent experiments (*P < 0.01).
To investigate the competition between the aptamer and hyaluronic acid in binding to HABD of CD44, we examined a combination of CD44 aptamer treatment with Low Molecular Weight (LMW-HA) of small hyaluronic at a concentration of 1 mg.mL-1 in the MDA-MB-231 cell. This concentration was in the range used by other studies, where cultured tumor cells were exposed to HA (12). LMW-HA was used because tumor microenvironment hyaluronidases degrade hyaluronic acid to smaller fragments (13). The result of this experiment indicated that snail expression can significantly decrease after combined treatment of LMV-HA and aptamer. Although combined treatment decreased the snail expression level, more reduction in snail expression was observed in cells treated with aptamer only.
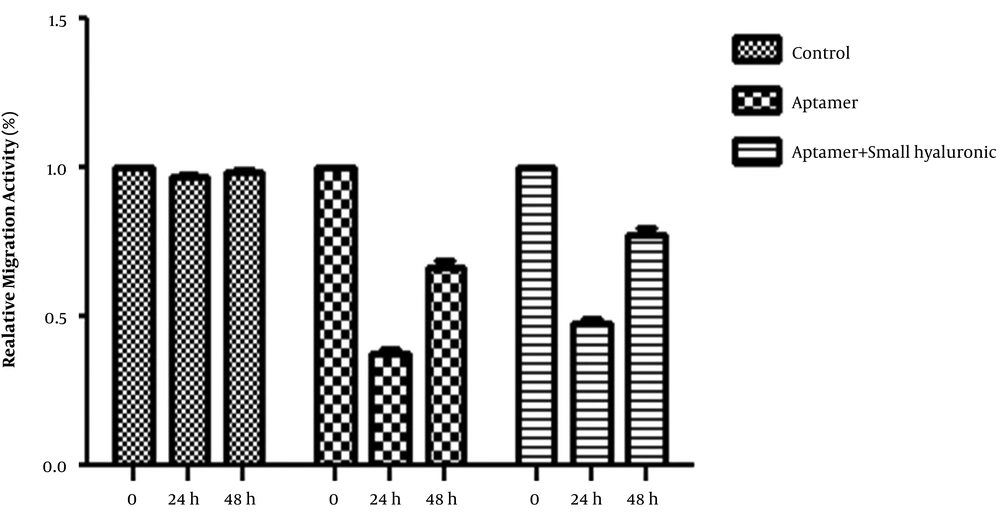
4.3. Aptamer Effect on Migration Ability of MDA-MB-231
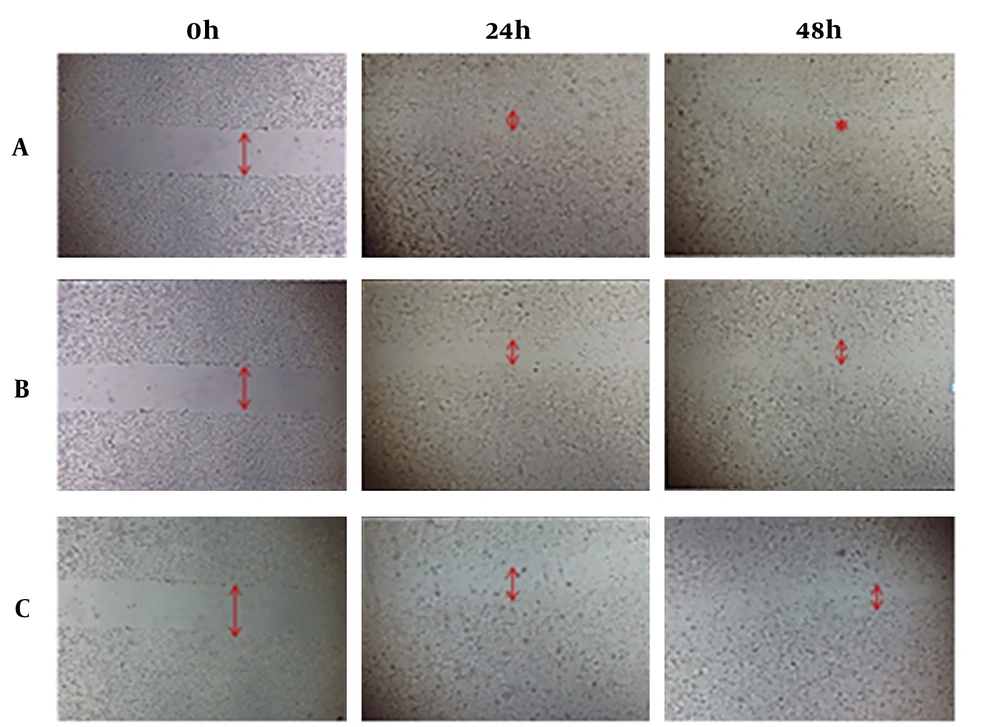
The effect of CD44 aptamer (50 ng.mL-1) alone, and in combination with LMW-HA (1 mg.mL-1) were examined on cell motility, using a wound healing assay at different time periods of 24 and 48 h. The gap was filled 30 h post-scratch in the negative control group, while 60% of the starting gap size was filled in the treated cell group in the same period. The distances between the border of the wound were determined 24 and 48 h post-scratching and compared with the initial measurement (0 h post-scratching), which were considered 100 %. The distances 24 h and 48 h after cell treatment with aptamer were 61 and 26 percent, respectively, whereas after treatment with a combination of aptamer and small hyaluronic acid were 51 and 22 percent, respectively. Our results confirmed that the migration potency of MDA-MB-231 cells in the aptamer-treated group was significantly lower in comparison with the control group. No significant difference was found between aptamer-treated and aptamer + HA-treated groups. The gap was completely filled 24 h post-scratching in the negative control group while in the treated group, about half of the initial gap size was filled. In the treated group, the gap was filled completely at about 50 h after scratching (Figures 4 and 5).
Cell migration and invasion assay. MDA-MB-231 control cells (A) and MDA-MB-231 cells treated with aptamer (B) and aptamer in combination with hyaluronic acid (C) were subjected to a wound-healing assay. Representative photographs at the indicated time points, from three independent experiments, each performed in triplicate that only one experiment is shown. Magnification: ×4. The amount of wound recovery was determined as described under materials and methods. The suppressing effects of the treatment on the migration ability at 0, 24 and 48 h post-scratching are shown.
5. Discussion
Mounting evidence indicates that interaction between HA and its major cell surface receptor, CD44 (14) can lead to intracellular signaling pathways stimulation, facilitating changes in proliferation, cell morphology, invasion and migration (15). These special effects on cellular behavior play a central role during normal development and cancer progression process. Independent studies showed a connection between the CD44+ phenotype of breast cancer cells and elevated expression of genes, which are involved in the invasion and the metastatic states. Although a direct relationship between snail expression and tumor metastasis has not been elucidated yet, a number of studies have exhibited that the overexpression of snail is associated with tumor invasion. In an effort to better understanding how CD44/HA inhibition might change snail expression pattern and consequently breast cancer invasion, we examined an aptamer that was developed based on Somasunderam et al. method (16), which specifically binds to CD44’s HABD with high affinity. Then, its inhibitory effect was investigated on snail expression in breast cancer cell line.
The result of this study indicated that inhibition of CD44/HA interaction by CD44 aptamer significantly reduced snail expression and aggressive behavior of MDA-MB231, according to the wound healing assay. The human MDA-MB231 breast cancer cell line was chosen due to its aggressive phenotype and high level of CD44 expression. These results seem to be in line with those of previous studies, indicating that CD44 knockdown repressed the ERK/Snail pathway in hepatocellular carcinoma (HCC) (17). Furthermore, CD44 knockdown by shRNA reduced cell invasion in 24 h as it was estimated by matrigel invasion assay (8). Inhibition of CD44 in human bladder cancer (14) and T24 cells repressed migration, angiogenesis, and invasion of these cells.
The real-time PCR data of aptamer-treated cell line suggest that a positive correlation can be detected between CD44/HA interaction and snail expression. Consistent with earlier findings, the results revealed that inhibition of CD44/HA interaction can reduce the snail expression and invasive behavior in the breast cancer cell line with a high level of CD44 expression as shown in Figures 2 and 3.
Previous studies indicated that CD44/HA interaction activates signaling pathways such as growth factor receptor signaling PI3/Akt and mitogen-associated protein kinase (MAPK), which promotes invasion (18). In addition, it was reported that PI3/Akt activates NF-κB through direct phosphorylation of IκB kinase α (IKKα). The NF-κB activation as one of the snail expression inducers leads to upregulation of snail (19). The MAPK signaling is another pathway, which is activated by CD44/HA interaction, and regulates induction of snail expression, and consequently EMT in collaboration with transforming growth factor-beta (TGF-β). Therefore, we hypothesized that inhibition of CD44/HA interaction might inactivate signaling of PI3/Akt and NF-κB by further reducing snail expression. Since snail plays a vital role in tumor growth and cancer metastasis (20, 21), it seems that the inhibitory effect of CD44/HA interaction reduces aggressive behavior of the treated cancer cell line (Figures 3 and 4).
In the group that was simultaneously treated with aptamer and small hyaluronic, the presence of HA did not affect the binding affinity of the aptamer. Snail expression and aggressive behavior were reduced in the combined treatment group; however, cells treated with only aptamer showed a reduction in the snail expression, which was more than those treated with aptamer plus HA. A possible explanation for this event might be due to HA reaction with cell-independent CD44 that can induce aggressive behavior. As previously stated, HA reacts to other cell receptors, except for CD44; therefore, HA might reduce the aptamer effect (6).
Many previous studies investigated the influence of anti-cancer agents and drugs in the immortalized cells that were cultured in 2D condition. Recently, investigations at in-vivo-like systems attracted much attention due to its similarity to tumor conditions in comparison with 2D culture system (5). The cells cultured in 2D monolayers or 3D cultures (in-vivo-like systems) exhibited different protein expression profiles and involved in signal transduction, cell-to-cell signaling, cellular movement, morphology, and cellular growth (12, 13). The present study indicates that cells cultured in 3D were more sensitive to aptamer treatment compared to the 2D model because the reduction in snail expression was more detectable. This difference can be explained by hypoxia, which activates hyaluronidase and stimulates HA production, ROS elevation and cell regulator of low oxygen tension, such as Hypoxia-inducible Factor 1α, β (HIF-1α, β). The HIF-1α regulates the CD44 expression and increases the level of CD44 molecules on the cell surface (22). These results suggest that cells in 3D model induce snail expression, leading to more sensitivity toward the suppressive effect of CD44 thioaptamer.
The use of aptamer in bed is increasing. Pegaptanib is the first FDA approved aptamer used in the treatment of Age-related Macular Degeneration (AMD) (23). Recently, Ulrich’s group conducted other studies to develop the application of their designed aptamer against CD126 to in vivo studies and clinical trials by modifying the aptamer to improve its resistannce against nucleases (24). One another clinical use of anti-CD aptamer-targeted delivery is in the generation of new vaccines. Wengerter et al. designed a modified RNA aptamer that could specifically target the overexpressed CD205 or DEC205 on dendritic cell surfaces. Administration of the anti-DEC205 aptamer-ovalbumin conjugates concomitant with adjuvant polyinosinic: polycytidylic acid (pIC), could promote T cell activation in vitro significantly (25). Currently, aptamers have become a powerful therapeutic and theranostic tool and demonstrated their abilities in several different clinical applications, such as cell phenotyping, immune modulation, cell-targeted delivery, and vaccine adjuvants. Besides, the clinical application of aptamers in other US Food and Drug Administration (FDA)-approved drugs and a handful of diagnostic kits for some random conditions have confirmed their successful and widespread applications in clinic (26). Given the extended clinical use of aptamers and our obtained results, the CD44 aptamer is a potential therapeutic target in cancer patients for instance breast cancer with a high level of CD44 expression.
5.1. Conclusion
In conclusion, the results of our study suggest that inhibition of HA-induced signaling is effective in reducing the invasive behavior of breast cancer cell lines. Furthermore, CD44 aptamer can be an effective agent in inhibiting metastasis and invasive behavior of aggressive breast cancer cell line with a high level of CD44 expression. These results can be useful for clinical use in the near future.