1. Background
Hepatitis B virus (HBV) infection is a serious global health problem and a significant cause of morbidity and mortality due to severe complications such as liver decompensation, cirrhosis, and hepatocellular carcinoma (HCC). Approximately one-third of all liver cancer deaths worldwide are attributed to HBV infection. Each year, about 820,000 people die from liver cirrhosis and HCC as a consequence of over 1.5 million new HBV infections annually (1, 2). The HBV establishes covalently closed circular DNA (cccDNA) for long-term persistence in hepatocytes (3). Chronic HBV infection persistence in the liver is linked to oxidative DNA damage, genomic instability, immune-mediated necro-inflammation, and cancer development (4).
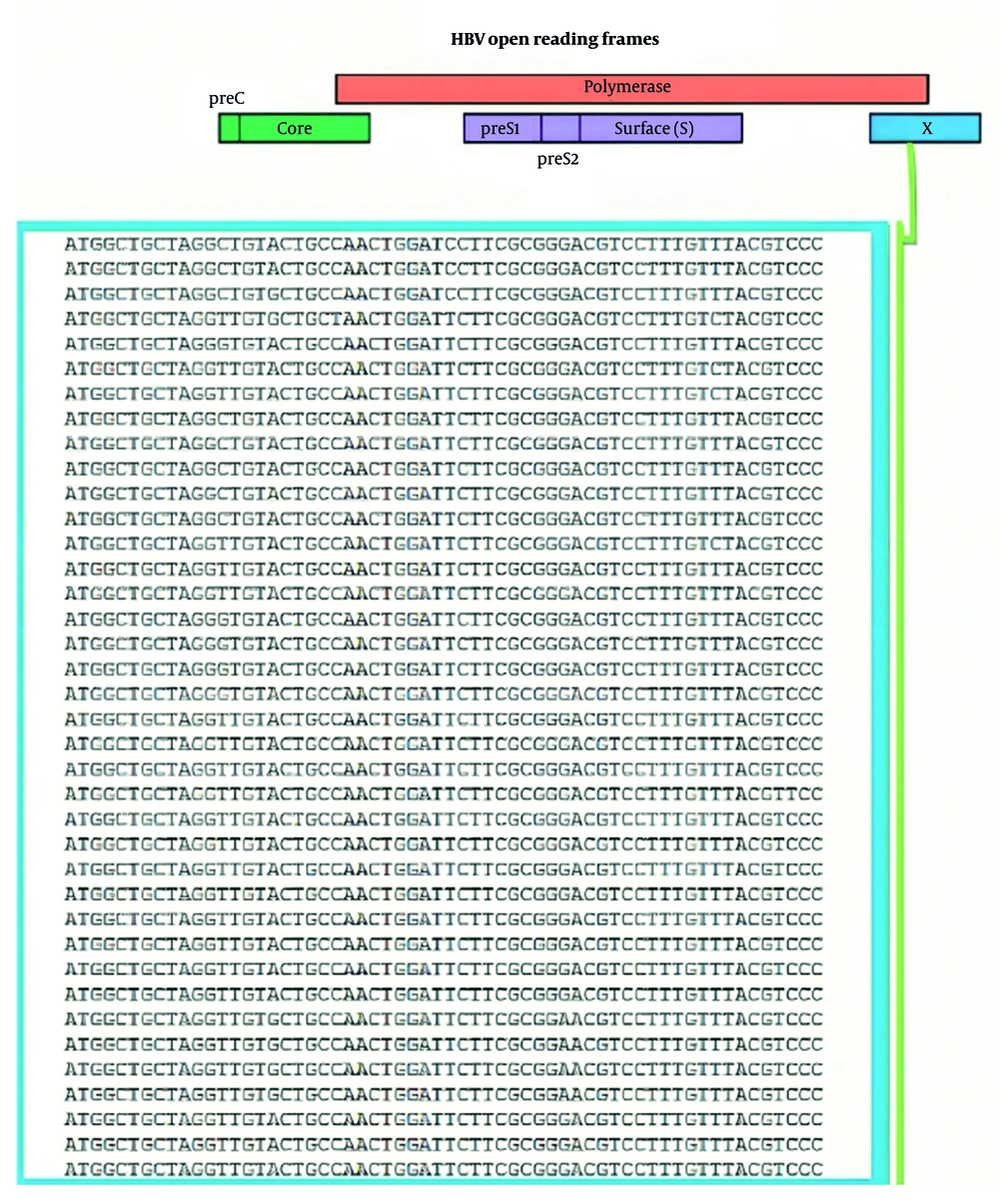
The HBV virions contain small (~3.2 kb) relaxed circular DNA (rcDNA), which converts to nuclear cccDNA. This cccDNA persists in hepatocyte nuclei, acting as a transcriptional mini-chromosome template for viral protein expression and genome replication (5). The genome consists of four overlapping open reading frames (ORFs) encoding preS/S, preC/C, P, and X proteins (6). The hepatitis B virus X protein (HBx) activates various viral and cellular promoters and enhancers and plays a central role in HBV-related hepatocarcinogenesis (7). The HBx participates in viral replication and promotes HCC via multiple mechanisms. It integrates into the hepatocyte genome, induces oxidative stress, activates oncogenic pathways, and drives epigenetic alterations disabling tumor suppressors and genomic stability. The HBx also enhances angiogenesis, extracellular matrix degradation, and metastasis by upregulating angiogenic factors, matrix metalloproteinases, and dysregulating cancer-associated microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), facilitating tumor progression (8).
RNA interference (RNAi) using short hairpin RNA (shRNA) or short interfering RNA (siRNA) offers a promising therapeutic approach for cancer (9). The RNAi achieves sequence-specific post-transcriptional gene silencing through double-stranded RNAs (10). It is initiated by 20 - 25 base pairs siRNAs or miRNAs, leading to efficient gene silencing. The siRNAs can also be expressed as shRNA from plasmid DNA (pDNA) or viral vectors under RNA polymerase III promoters (11). After transcription, shRNA moves to the cytoplasm for processing by Dicer into siRNA duplexes. Like synthetic siRNA, this endogenous siRNA binds target messenger RNA (mRNA) and loads into the RNA-induced silencing complex (RISC), causing mRNA degradation. Studies confirm RNAi’s therapeutic potential for diseases such as viral hepatitis and cancers (12).
The shRNAs are often more efficient than siRNAs for gene silencing due to continuous transcription within host cells, ensuring sustained siRNA supply after Dicer processing. Synthetic siRNAs, however, are transient and are gradually degraded or diluted during cell division, resulting in shorter silencing duration (13). A key shRNA advantage over siRNAs is stable genomic integration via viral vectors, providing durable knockdown (14). The RNAi-based therapies targeting HBV offer viral gene specificity and can inhibit HBV replication and integrated viral DNA transcripts, potentially reducing side effects versus conventional treatments. Simultaneously targeting multiple viral regions may enhance efficacy and lower risks of disease progression and HCC development (15).
2. Objectives
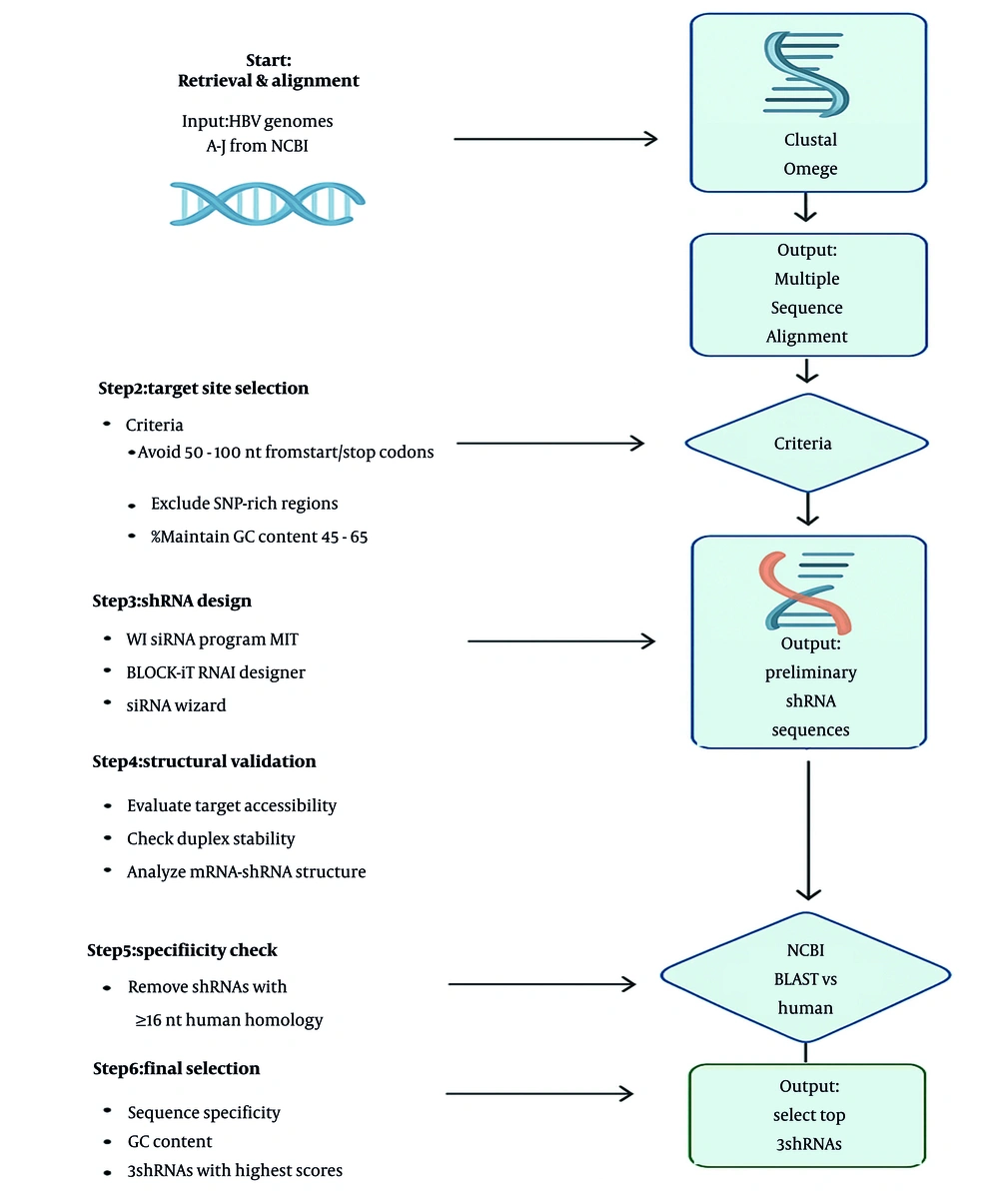
The current research focused on the computational design of potential shRNAs targeting the hepatitis B virus X (HBX) gene of HBV to provide a foundation for future therapeutic development.
3. Methods
3.1. Selection of Hepatitis B Virus X Gene Conserved Regions
Based on the genome sequence, HBV has been classified into 10 well-known genotypes (A to J) (16). The major classification of HBV genotypes by sequence divergence has been further divided into subgenotypes or serotypes. Based on serological reactivities of hepatitis B surface antigen (HBsAg), HBV is classified into four major subtypes: The adw, adr, ayw, and ayr (17). Generally, adw subtypes occur in all genotypes except D and E, while ayw subtypes dominate genotypes B and D. Conversely, adr and ayr subtypes associate primarily with genotype C (18, 19).
Complete HBV reference sequences were obtained from the viral gene bank database within the National Institutes of Health (NIH) genetic sequence database (20). The Clustal Omega website — a multiple sequence alignment tool — was used to align complete coding sequences (CDS) and select conserved regions (21). For shRNA design, the HBX region of HBV genotypes was targeted. Note that mismatches between target mRNA and shRNA significantly affect shRNA efficacy. This approach ensures engineered shRNA molecules correspond to conserved consensus sequences. Consequently, subsequent analysis focused on regions of high homology between target sites and mRNAs.
3.2. Development of Short Hairpin RNA Molecules Targeting the Hepatitis B Virus X Gene
Three online tools were used to design shRNA: The WI siRNA Selection Program, Invitrogen BLOCK-iT RNAi Designer (https://rnaidesigner.lifetechnologies.com/rnaiexpress/), and siRNA Wizard Software (InvivoGen) (22). Each tool offers distinct advantages, with parameters and siRNA activity features calculated based on application (in vitro, in vivo) and targets (cellular, viral) (23).
For improved shRNA design, this study carefully evaluated guide structure and sequence characteristics. Sequence selection should avoid regions within 50 - 100 nucleotides of start/stop codons due to transcriptional factor occupancy, while also excluding single nucleotide polymorphism (SNP) sites and intronic SNPs. Target sequence GC content must be 45 - 65% for shRNA stability (24, 25). Structural rules address thermodynamic and secondary structure features of the target site, defining binding energy functionality and optimal duplex energy differentials. The mRNAs lacking rigid secondary structures exhibit stronger shRNA binding than highly structured conformations. These rules control shRNA target accessibility and shRNA/target duplex thermodynamics (26).
CLC Genomics Workbench software predicted secondary structures of the HBX gene and designed shRNAs interactions (14). Constructed sequences underwent manual analysis using algorithms by Ui-Tei et al., Reynold et al., and Jagla et al. to optimize shRNA efficiency (27-29).
3.3. Similarity Search
To evaluate the specificity of the designed shRNAs, a Nucleotide BLAST search was performed using the human genomic and transcript database to identify off-target sequence similarities in non-targeted genomes (22). Based on the results, shRNAs exhibiting 16 or more consecutive nucleotides of homology to any other mRNAs were excluded from the suggested list (24, 30).
3.4. Secondary Structure Prediction of Hepatitis B Virus X Gene
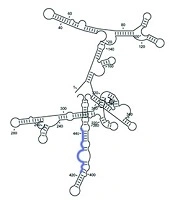
The GC content of the predicted shRNAs was calculated using the Oligo Analyzer Calculator (31). GC content influences shRNA stability, functionality, and target affinity. Higher GC content may slow duplex unwinding, while lower GC percentage can compromise target affinity efficiency. Secondary structures of HBX were predicted using CLC Genomics Workbench software. To verify shRNAs alignment with target gene regions, a BLAST search was conducted using the predicted mRNA secondary structure. The shRNAs located in stem or congested areas were excluded from analysis.
3.5. Choosing Short Hairpin RNAs with the Best Score
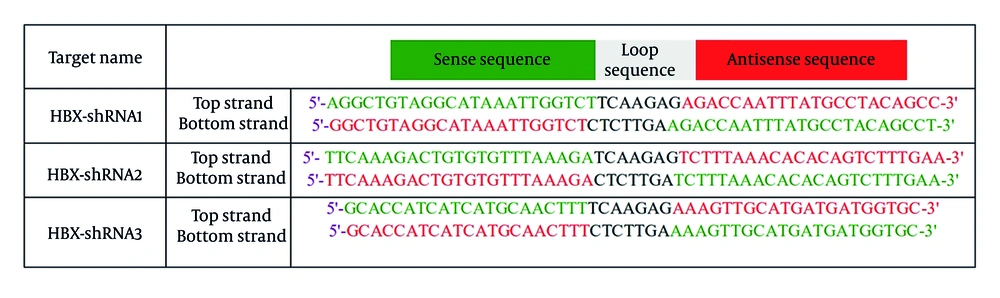
In this study, the sequence specificity, GC content, and off-target potential of the predicted shRNAs were reevaluated to verify their effectiveness. According to the results, the three highest-scoring shRNAs were ultimately chosen. The nucleotide sequence TCAAGAG served as the loop sequence between the sense and antisense strands (14).
4. Results
4.1. Sequence Alignment
The methodology of RNAi is applicable for targeting viral genes with high degrees of sequence conservation. In this study, approximately 100 gene sequences from different HBV genotypes were obtained from the NCBI GenBank database. The Clustal Omega sequence alignment program identifies consensus regions through multiple sequence alignment. The HBX conserved regions were considered for shRNA selection (Figure 1).
4.2. Short Hairpin RNA Design
The BLOCK-iT RNAi Designer, WI siRNA Selection Program, and siRNA Wizard online tool were utilized to provide functional shRNA designs. These resources were carefully examined to design the most efficient molecules. The WI siRNA Selection Program effectively demonstrated the off-target effects associated with the siRNA guide strand's seed region and complementary sequences in target sites across various species. The Whitehead server proposed sixteen shRNAs, BLOCK-iT RNAi Designer proposed ten, and Invitrogen proposed eight; these were manually reviewed for optimal design. The numbers of shRNAs meeting the desired parameters are listed in Table 1. Before final scoring, BLAST searches were performed against expressed sequences to ensure the shRNA constructs targeted only HBX.
| HBX shRNA ID | Location of Target Within mRNA b | Sequence | Length | GC % c | Conserved Sequence d | Un-conserved Sequence e | U at Position 10 (Sense) f | Proper Regions of the RNA Secondary Structure g | Crowded Regions of the RNA Secondary Structure h |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 266 | TGCCCAAGGTCTTACATAAGAGG | 23 | 42 | * | - | * | - | * |
| 2 | 334 | TTCAAAGACTGTGTGTTTAAAGA | 23 | 32 | * | - | * | * | - |
| 3 | 422 | GCACCATCATCATGCAACTTT | 21 | 42.86 | * | - | * | * | - |
| 4 | 273 | GGTCTTACATAAGAGGACTCTTGGA | 25 | 44 | - | * | * | * | - |
| 5 | 284 | AGAGGACTCTTGGACTCCCAGCAAT | 25 | 52 | * | - | * | * | - |
| 6 | 404 | AGGCATAAATTGGTCTGCGCACCAT | 25 | 48 | * | - | * | * | - |
| 7 | 393 | TAGGAGGCTGTAGGCATAAATTG | 23 | 47 | * | - | - | * | - |
| 8 | 397 | AGGCTGTAGGCATAAATTGGTCT | 23 | 42 | * | - | - | * | - |
| 9 | 338 | AAGACTGTGTGTTTAAAGACTGG | 23 | 37 | - | * | * | - | * |
Abbreviations: HBX, hepatitis B virus X gene; shRNA, short hairpin RNA; mRNA, messenger RNA.
a In the table constructed based on the scoring system, each shRNA that met the specified criteria is marked with an asterisk (*), and ultimately, the sequences with the highest overall scores are selected.
b Location of target within mRNA (locus on the HBX gene).
c GC content (should be in the range of 45 - 65%).
d Conserved sequence [conserved regions among the hepatitis B virus (HBV) genotypes].
e Un-conserved sequence (un-conserved regions among the all genotypes of HBV).
f U at position 10 (sense) potentially improving RISC cleavage.
g Proper regions of the RNA secondary structure (accessible and unstructured regions of the target mRNA that allow efficient binding of the shRNA).
h Crowded regions of the RNA secondary structure (these are highly structured or sterically hindered parts of the mRNA that are less suitable for shRNA binding).
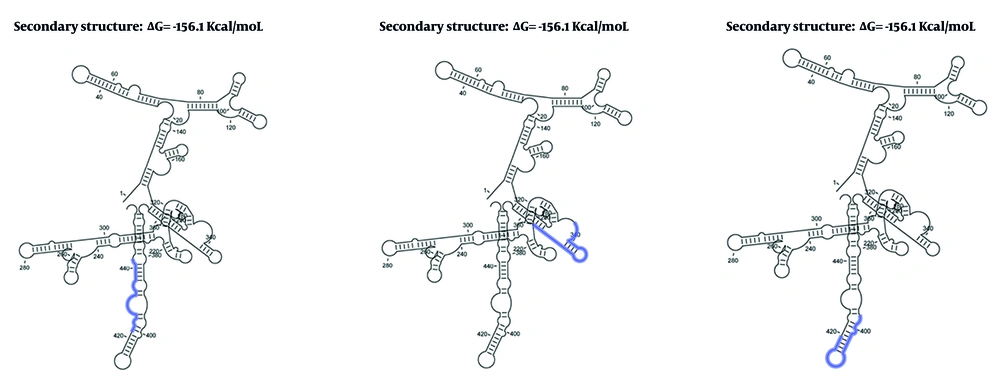
4.3. Secondary Structure and Interactions
CLC Genomics Workbench was applied to predict the RNA secondary structure and interactions between shRNAs and mRNA, as shown in Figure 2. We determined that HBX shRNAs 1, 2, and 3 were highly potent in reducing viral mRNA and protein expression (Figure 3). The shRNA design workflow is schematically summarized in Figure 4.
5. Discussion
The application of shRNA technologies presents several advantages, including the stable integration of expression constructs into genomic DNA to support prolonged expression. Furthermore, viral vectors can infect typically hard-to-target cell lines and tissues, while shRNA transcription can be temporally regulated using inducible promoters (13). In HBV-infected patients, HBx is frequently expressed in HCC tissue, where it facilitates the activation of various viral and cellular promoters and enhancers crucial for viral replication and HCC development. The HBx coding region integrates into specific sites within the host cell's chromosomal DNA (32). Additionally, HBx expression regulates numerous cellular signal transduction pathways involved in cell migration and invasion (33). Knockdown of the HBX gene by shRNAs disturbs these critical functions.
Despite considerable progress in controlling and preventing many viral diseases, the lack of effective drugs against most viral infections remains a major medical need. Therefore, the clinical application of RNAi is important in gene therapy because it allows precise targeting of viral genes. For example, Huang et al. used DNA vector-based shRNAs to prevent influenza virus infection in vitro by targeting the PB2 gene (34). Cheng et al. conducted a study to inhibit HBsAg expression in HBV models using a shRNA expression system (35). Ter Brake’s team tested a strategy employing multiple shRNAs against the human immunodeficiency virus type 1 (HIV-1) Pol and Gag genes to avoid viral escape (36). However, shRNA molecules offer advantages over siRNA, including compatibility with viral vectors; therefore, we applied shRNA molecules to prevent HBV replication by repressing the HBX gene.
We designed these molecules using three online tools: BLOCK-iT RNAi Designer, WI siRNA Selection Program, and siRNA Wizard. We predicted the secondary structures and target accessibility of the shRNA molecules using CLC Genomics Workbench software. To predict effective molecules, we diligently assessed scoring criteria such as GC content, an uracil (U) residue at position 10, DNA sequence conservation, BLASTN specificity analysis, and uncrowded regions in RNA secondary structures. Finally, we selected three molecules with the highest score. We plan to apply a lentivirus-mediated shRNA expression vector to prevent HBX gene expression in HCC cell line models.
An in-silico prediction and experimental validation of siRNAs targeting hepatitis B/C viruses has been performed previously (37, 38). He et al. indicated that cells with HBx knockdown showed increased sensitivity to 5-fluorouracil and cisplatin treatment (39). Moreover, combining HBx-targeted RNAi with chemotherapy significantly enhanced apoptosis and inhibited proliferation in HCC cells. The findings demonstrate that designed shRNA constructs effectively suppress HBx expression and consequently inhibit HBV replication in HCC cell lines, providing strong evidence for RNAi as a promising therapeutic strategy against HBV persistence and its oncogenic pathways.
Importantly, the present study offers a distinct advancement over previous HBV shRNA designs by applying a pan-genotypic sequence conservation analysis across HBV genotypes A - J to identify highly conserved domains within the HBX gene. This ensures broader antiviral coverage and significantly reduces the potential for viral escape mutations. Additionally, thermodynamic models and siRNA efficacy scoring algorithms were integrated to optimize shRNA stem-loop structures for enhanced potency and minimal off-target effects; host transcriptome screening further improved specificity and safety.
The designed shRNA constructs will be experimentally validated in HCC cell lines, such as HepG2, Hep3B, and Huh7, to assess their ability to suppress HBX gene expression and inhibit viral replication. The efficiency of these constructs in downregulating viral transcripts and proteins will be evaluated using quantitative reverse transcription polymerase chain reaction (RT-PCR) and Western blot analyses. Functional assays will measure their effects on cell viability, proliferation, and apoptosis. If successful, these findings could lead to the use of shRNA-mediated HBX silencing as a therapeutic strategy to suppress HBV infection and prevent HBx-associated hepatocarcinogenesis. This approach could contribute to developing novel RNAi-based gene therapies for chronic HBV infection and HBV-induced liver cancer.
5.1. Conclusions
In conclusion, a critical gap remains in the availability of effective antiviral agents for treating HBV infection. The shRNA therapeutics represent a promising strategy due to their capacity for sustained gene silencing and prolonged expression. By specifically targeting and inhibiting pathogenic viral genes, shRNA-based interventions could significantly reduce disease progression. This sustained antiviral effect may substantially decrease the burden of HBV infection, offering new hope for patients and healthcare providers in managing this widespread disease. In this study, we designed and computationally evaluated a series of shRNAs targeting the HBX gene as a potential RNAi-based therapeutic strategy. Structural and functional predictions suggest these shRNAs effectively bind and silence HBX transcripts, potentially reducing HBx protein levels and inhibiting viral replication and associated oncogenic pathways. Given HBx's central role in HBV persistence, oxidative stress induction, and HCC progression, silencing this gene may provide dual benefits by suppressing viral replication and limiting HBV-induced tumorigenesis. Future validation in HCC cell lines will determine their functional efficacy and therapeutic applicability.